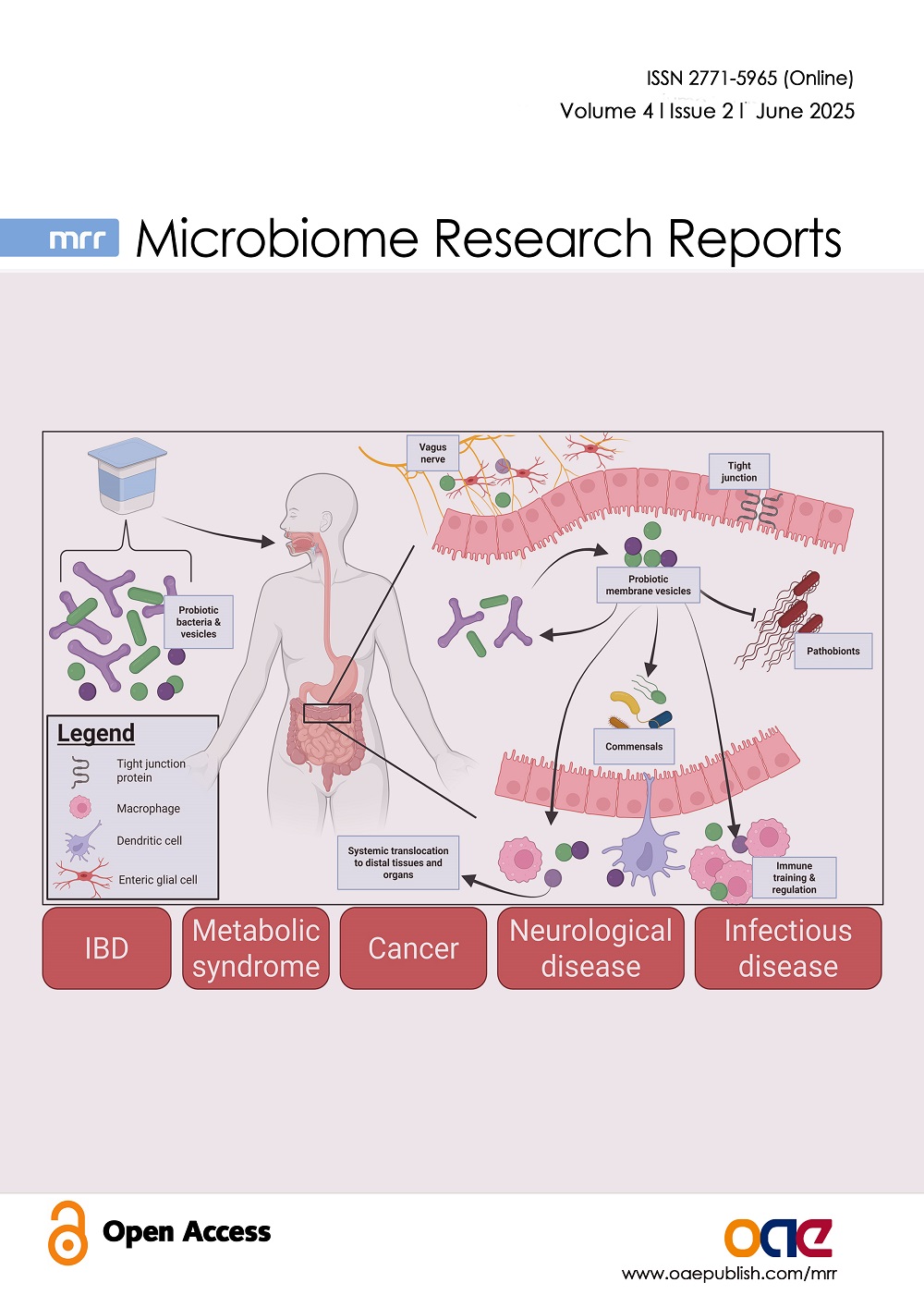
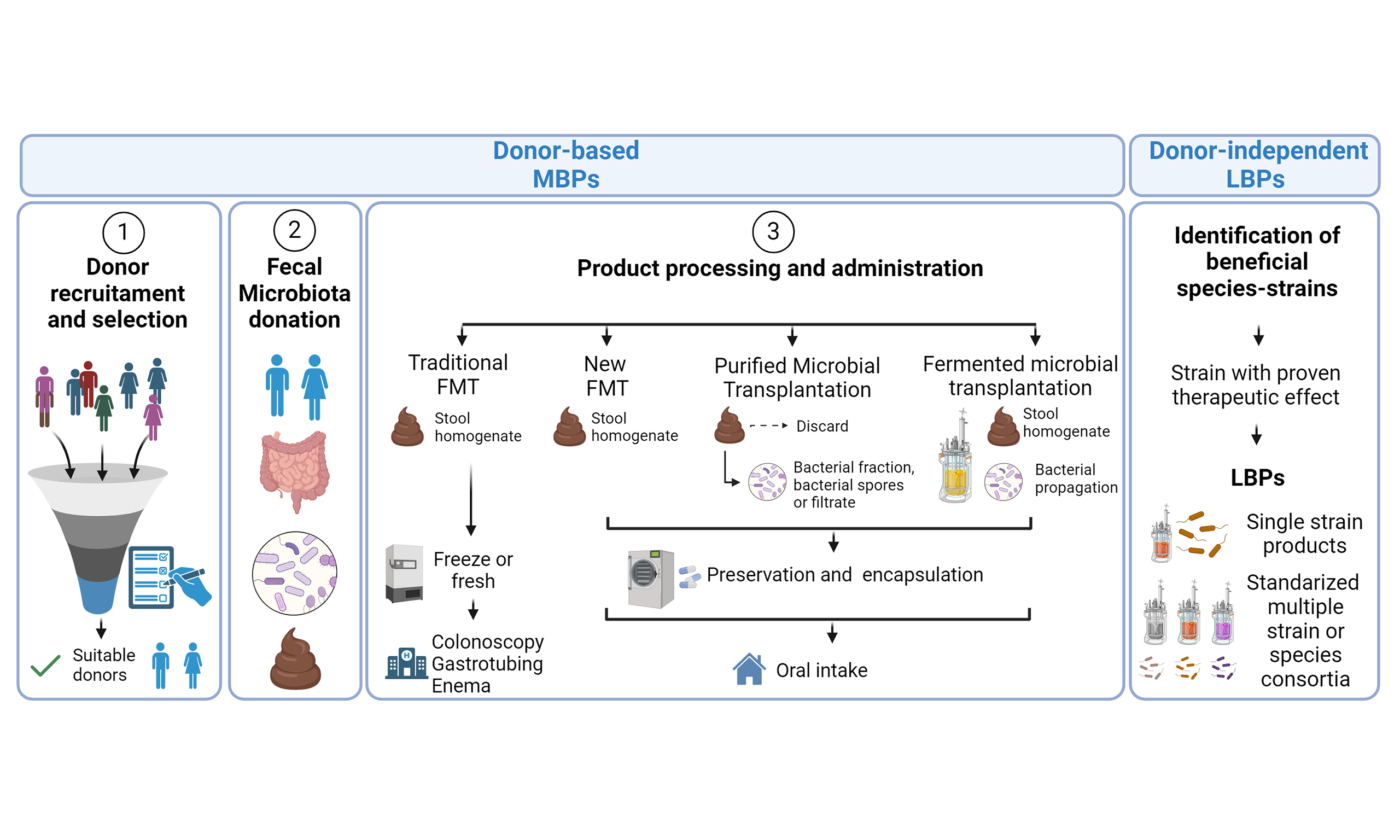
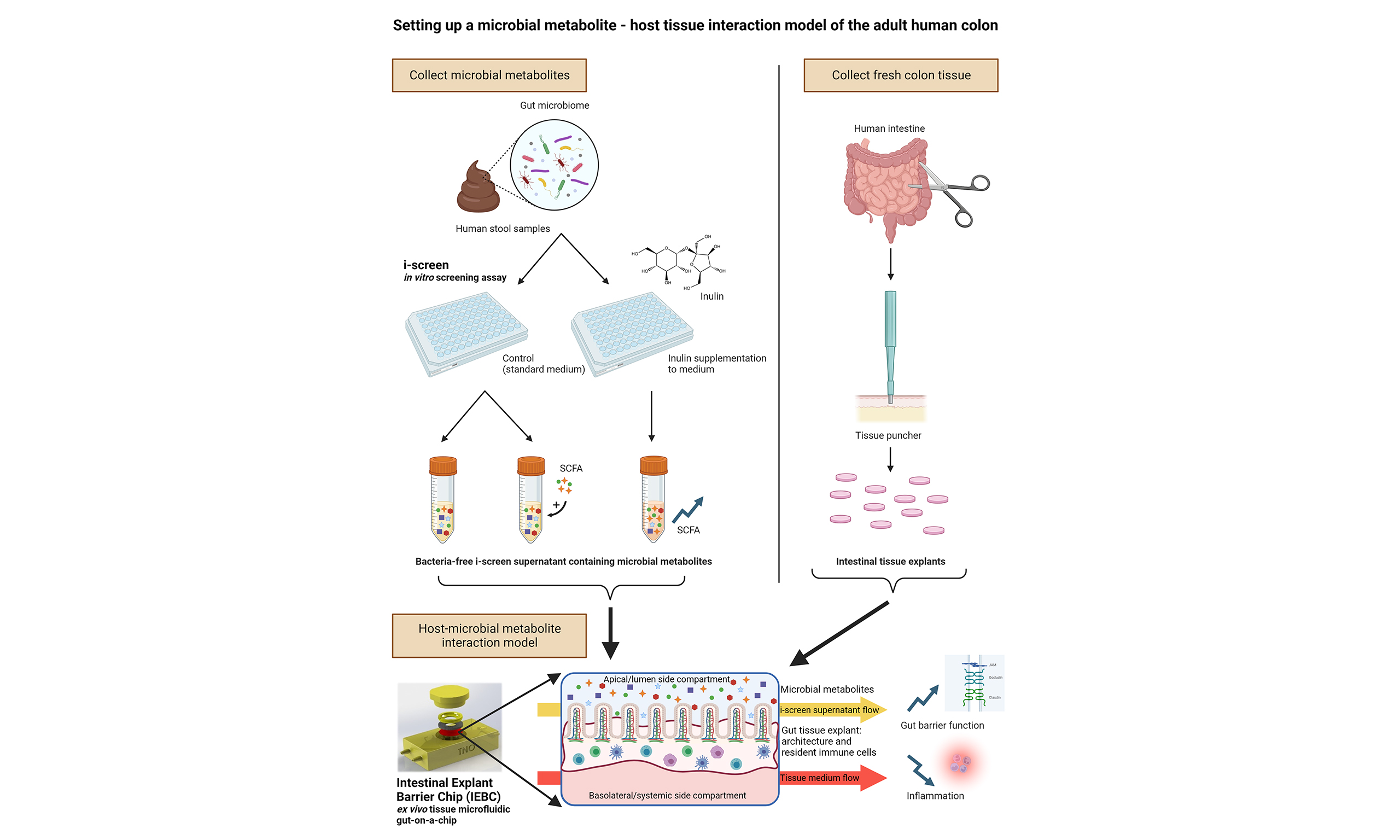
Microbiome Research Reports

Data
905
Authors
1490
Reviewers
2021
Published Since
For Reviewers
For Readers
Add your e-mail address to receive forthcoming Issues of this journal:
Themed Collections
Volume 4, Issue 4 (in progress)
Volume 4, Issue 3
Volume 4, Issue 2
Volume 4, Issue 1
Volume 3, Issue 4
Volume 3, Issue 3
Volume 3, Issue 2
Volume 3, Issue 1
Volume 2, Issue 4
Volume 2, Issue 3
Volume 2, Issue 2
Volume 2, Issue 1
Volume 1, Issue 4
Volume 1, Issue 3
Volume 1, Issue 2
Volume 1, Issue 1

Coming soon.
Volume 4, Issue 4 (in progress)
Volume 4, Issue 3
Volume 4, Issue 2
Volume 4, Issue 1
Volume 3, Issue 4
Volume 3, Issue 3
Volume 3, Issue 2
Volume 3, Issue 1
Volume 2, Issue 4
Volume 2, Issue 3
Volume 2, Issue 2
Volume 2, Issue 1
Volume 1, Issue 4
Volume 1, Issue 3
Volume 1, Issue 2
Volume 1, Issue 1
Data
905
Authors
1490
Reviewers
2021
Published Since