fig1
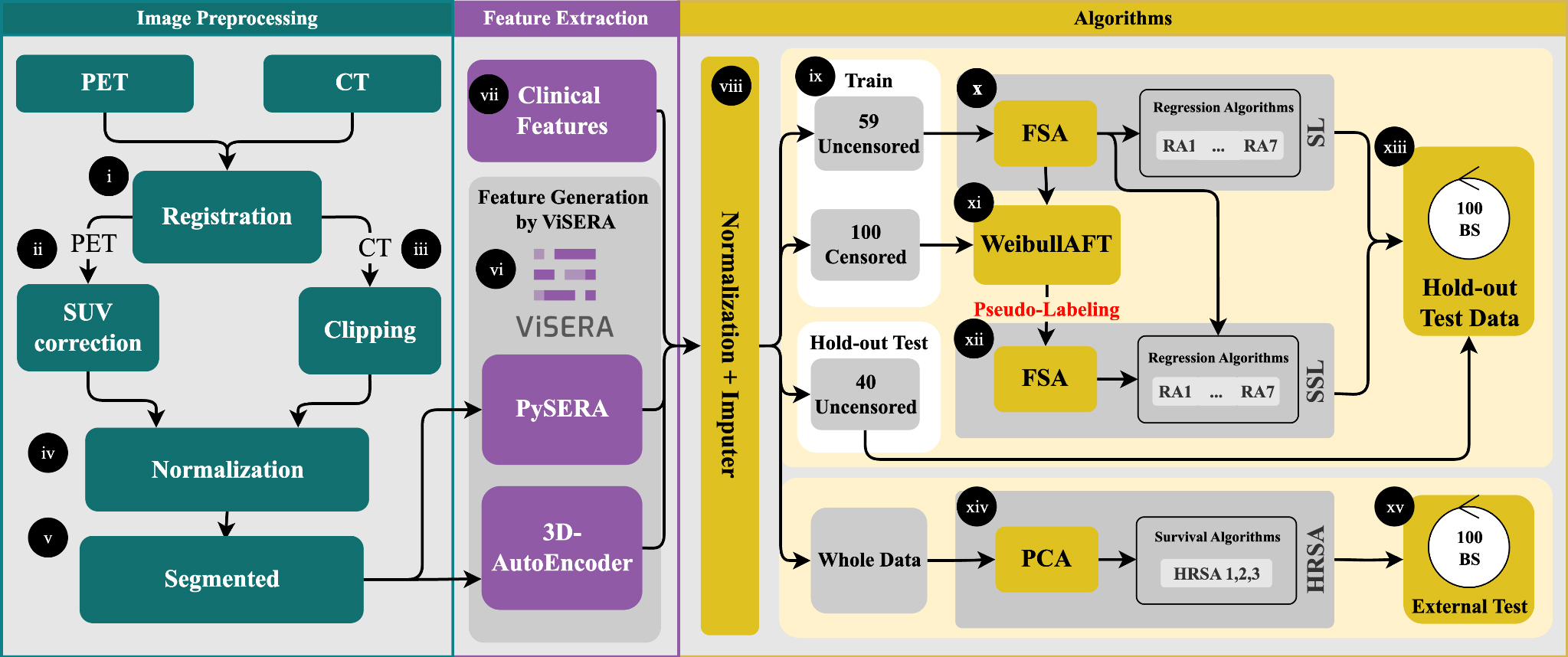
Figure 1. Preprocessing included PET-to-CT registration, SUV correction, CT clipping, and min-max normalization (i-iv). Tumor ROIs were delineated for feature extraction (v), yielding 215 handcrafted radiomics features (HRFs) and 1024 deep radiomics features (DRFs) (vi), which were combined with clinical features (CFs) to generate 13 datasets (vii). After dataset generation, features were normalized, missing values imputed, and the data split into an uncensored holdout set (40 cases) and censored vs. uncensored training sets (100/59 cases) (viii-ix). Feature selection (RR and FR) reduced dimensionality to 20 features, followed by training with seven regression algorithms (x-xii). Survival modeling used both supervised (SL) and semi-supervised (SSL) strategies, with pseudo-labeling applied to censored cases. Model performance was evaluated on the holdout set using MAE, C-index, log-rank tests, and SHAP-based feature importance (xiii-xiv). External validation was conducted on 33 TCIA patients (xv). PET: Positron emission tomography; CT: computed tomography; ROIs: regions of interest; RR: Pearson’s correlation coefficient regression; FR: F-test for regression; MAE: mean absolute error; SHAP: SHapley Additive exPlanations; TCIA: cancer imaging archive.