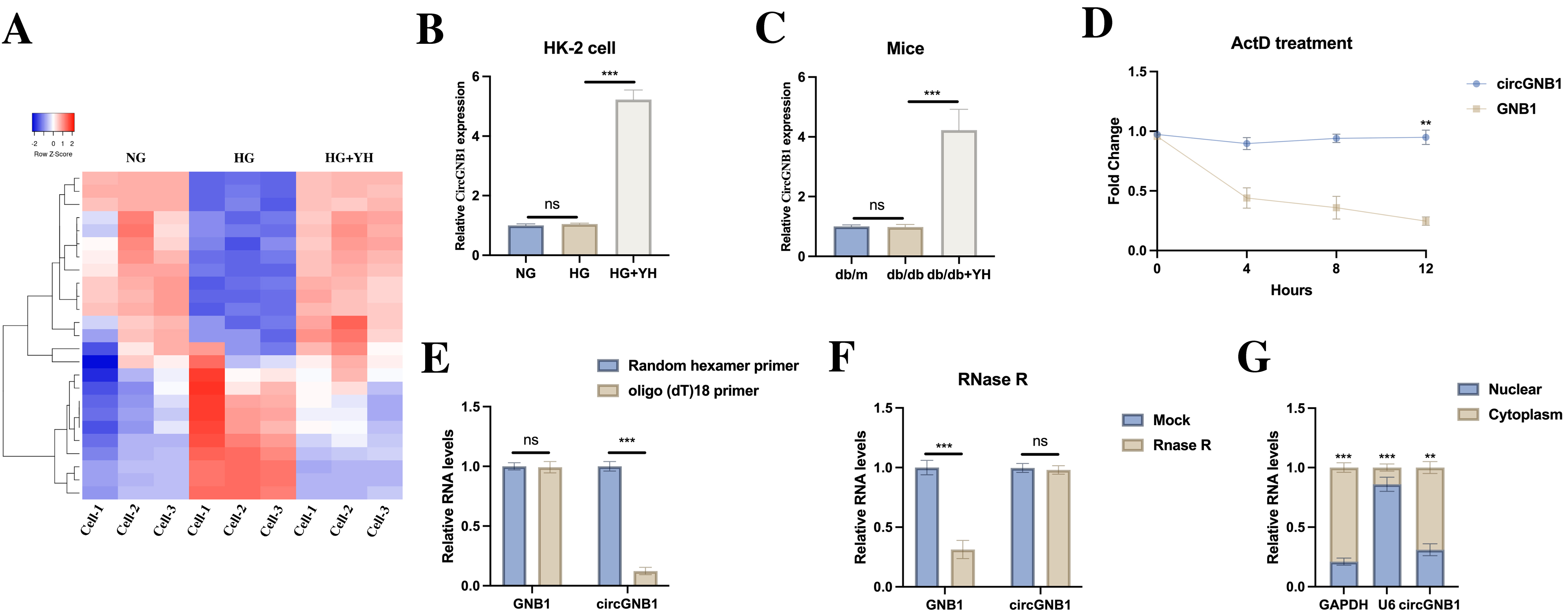
fig3
Figure 3. CircGNB1 upregulation by YH in vitro and in vivo. (A) CircRNA microarray analysis of differentially expressed circRNAs; (B) CircGNB1 expression in NG, HG, and YH-treated HG cells; (C) CircGNB1 levels in renal tissues from different treatment groups; (D) RNA stability assessment after actinomycin D treatment; (E) Reverse transcription efficiency using random hexamer versus oligo(dT)18 primers; (F) RNase R resistance test comparing circular and linear RNA forms; (G) Subcellular localization of circGNB1 in HK-2 cells determined by fractionation and FISH analysis (Scale bar = 20 μm). **P < 0.01; ***P < 0.001. HG: High glucose; YH: yohimbine; NG: normal glucose; circRNA: circular RNA; RNA-seq: RNA sequencing; RT: reverse transcription; FISH: fluorescence in situ hybridization; GNB1: guanine nucleotide-binding protein subunit beta-1; circGNB1: circular RNA derived from the GNB1 gene; GAPDH: glyceraldehyde-3-phosphate dehydrogenase; U6: U6 small nuclear RNA.