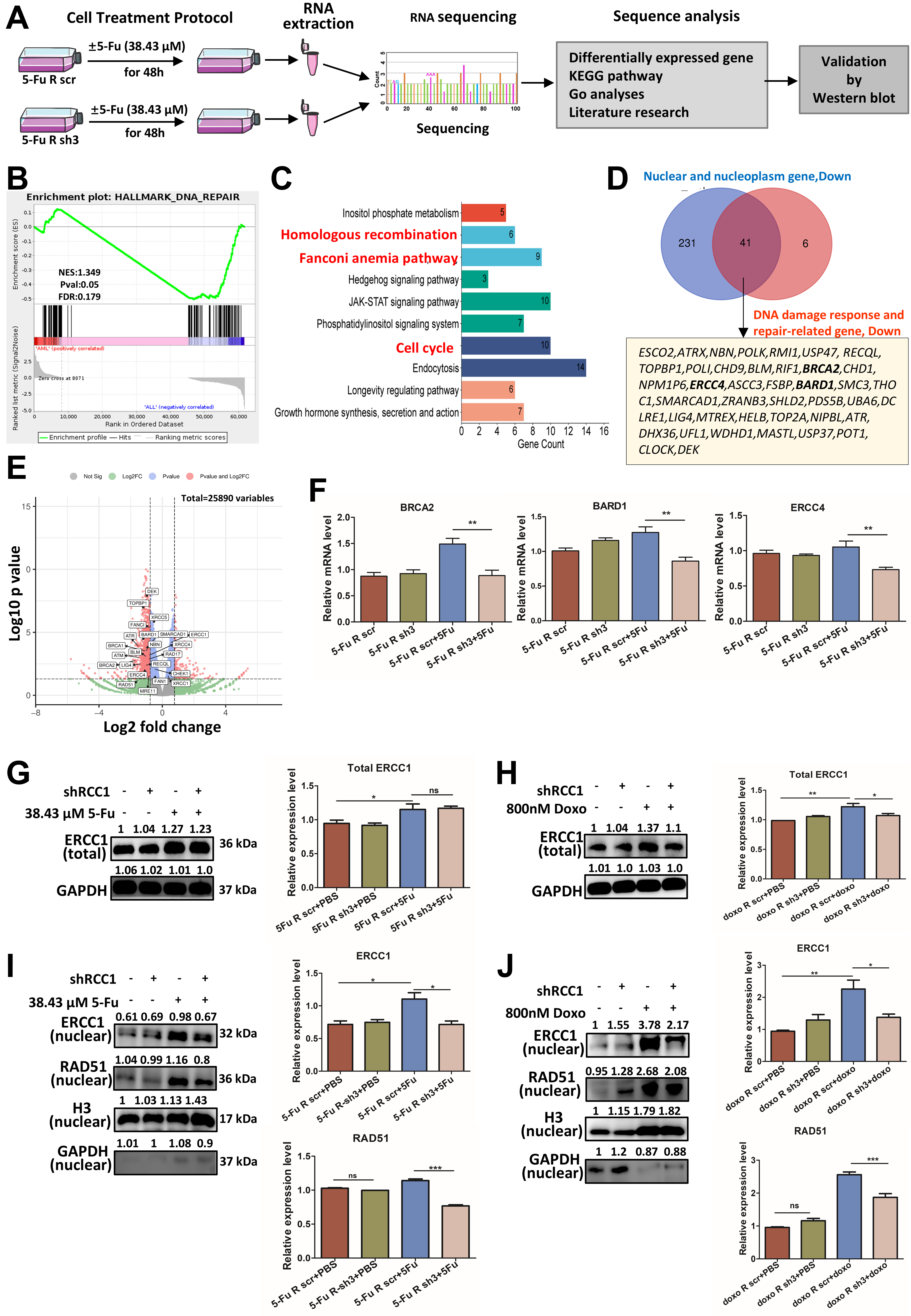
fig7
Figure 7. RCC1 knockdown disrupts the expression and nuclear localization of DNA repair proteins. (A) Schematic overview of the workflow used to investigate molecular changes in 5-FU-resistant HCT116 cells with or without RCC1 knockdown (n = 2 per group); (B) GSEA showing significant downregulation of DNA repair pathways in the RCC1 knockdown + 5-FU group; (C) KEGG pathway enrichment analysis of differentially expressed genes, highlighting downregulated pathways including HR, FA, and cell cycle pathways; (D) GO analysis focusing on nuclear and DNA repair-related genes identified 41 potential downregulated targets (fold change < 0.5, RCC1 knockdown + 5-FU vs. control + 5-FU); (E) Volcano plot of differentially expressed genes in the RCC1 knockdown plus 5-FU treatment group; DNA repair-related genes are highlighted; (F) qRT-PCR analysis of ERCC4, BRCA2, and BARD1 RNA levels, showing significant reductions upon RCC1 knockdown (one-way ANOVA, P < 0.01); (G and H) Western blot analysis of total ERCC1 protein levels (one-way ANOVA); GAPDH served as an internal control; (I and J) Western blot analysis of nuclear ERCC1 and RAD51 levels (one-way ANOVA); H3 was used as a loading control. ns, not significant; ***P < 0.001; **P < 0.01; *P < 0.05. RCC1: Regulator of chromosome condensation 1; 5-FU: 5-fluorouracil; GSEA: gene set enrichment analysis; KEGG: Kyoto Encyclopedia of Genes and Genomes; HR: homologous recombination; FA: Fanconi anemia; GO: Gene Ontology; qRT-PCR: quantitative reverse transcription-polymerase chain reaction; ANOVA: analysis of variance; GAPDH: glyceraldehyde-3-phosphate dehydrogenase.