fig1
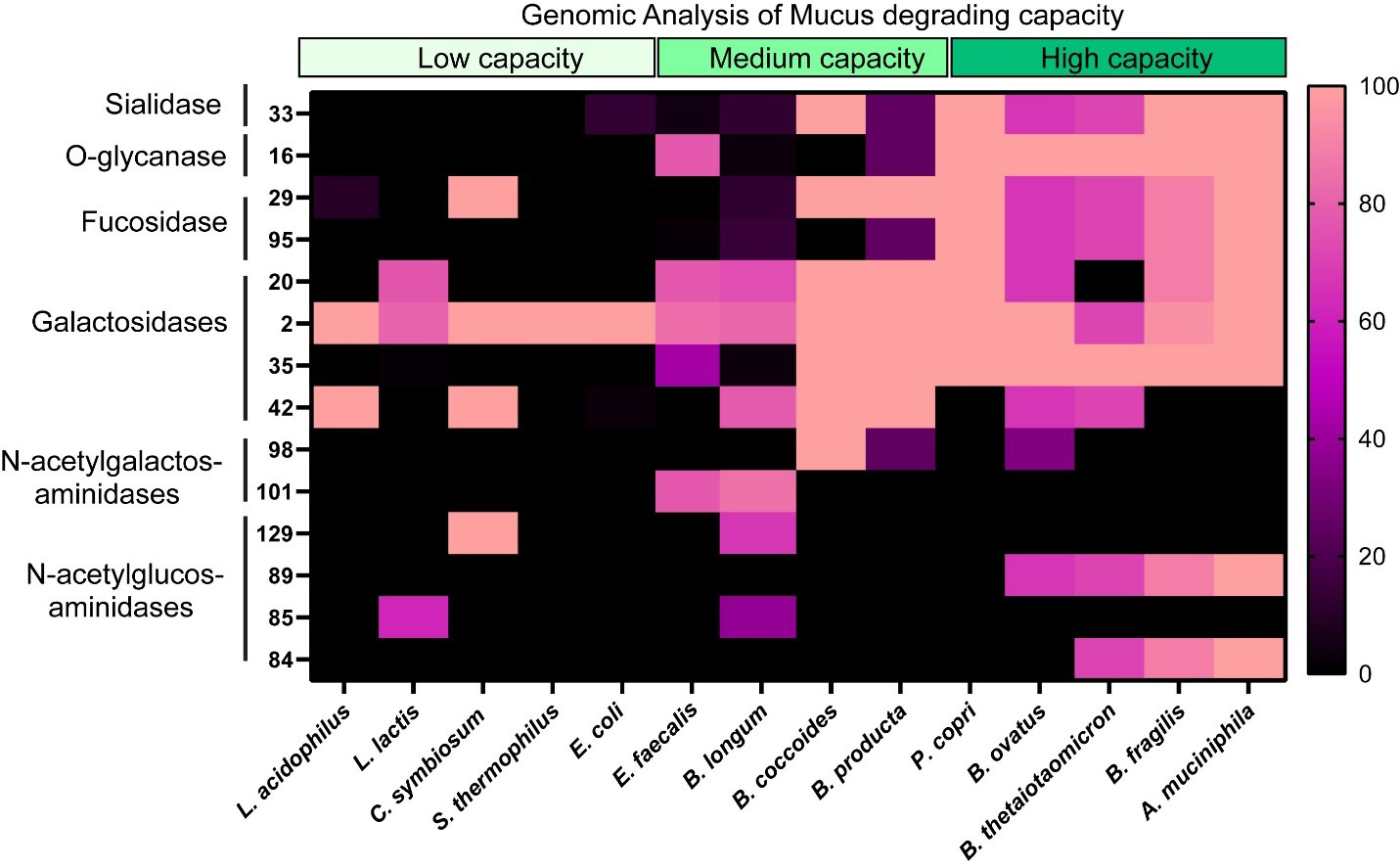
Figure 1. Heat map showing the percentage of bacterial genomes encoding mucin glycan-associated GHs. GH families examined included GH 33, 16, 29, 95, 20, 2, 35, 42, 98, 101, 129, 89, 85, and 84, all of which are involved in mucin glycan degradation. Genome counts and GH annotations were obtained from the CAZy database. Percentages were calculated as the number of genomes containing a given GH family divided by the total number of genomes analyzed for that taxon, multiplied by 100 (See Equation 1). Higher values shown in pink indicate a greater proportion of genomes encoding the specified GH. Genomes analyzed included: L. acidophilus, L. lactis, C. symbiosum, S. thermophilus, E. coli, E. feacalis, B. longum, B. coccoides, B. producta, P. copri, B. ovatus, B. thetaiotaomicron, B. fragilis, and A. muciniphila genomes. Graph generated using GraphPad Prism software. L. acidophilus: Lactobacillus acidophilus; L. lactis: Lactococcus lactis; C. symbiosum: Clostridium symbiosum; S. thermophilus: Streptococcus thermophilus; E. coli: Escherichia coli; E. feacalis: Enterococcus feacalis; B. longum: Bifidobacterium longum; B. coccoides: Blautia coccoides; B. producta: Blautia producta; P. copri: Prevotella copri; B. ovatus: Bacteroides ovatus; B. thetaiotaomicron: Bacteroides thetaiotaomicron; B. fragilis: Bacteroides fragilis; A. muciniphila: Akkermansia muciniphila; GH: glycosyl hydrolase; CAZy: Carbohydrate-Active enZYmes.