fig1
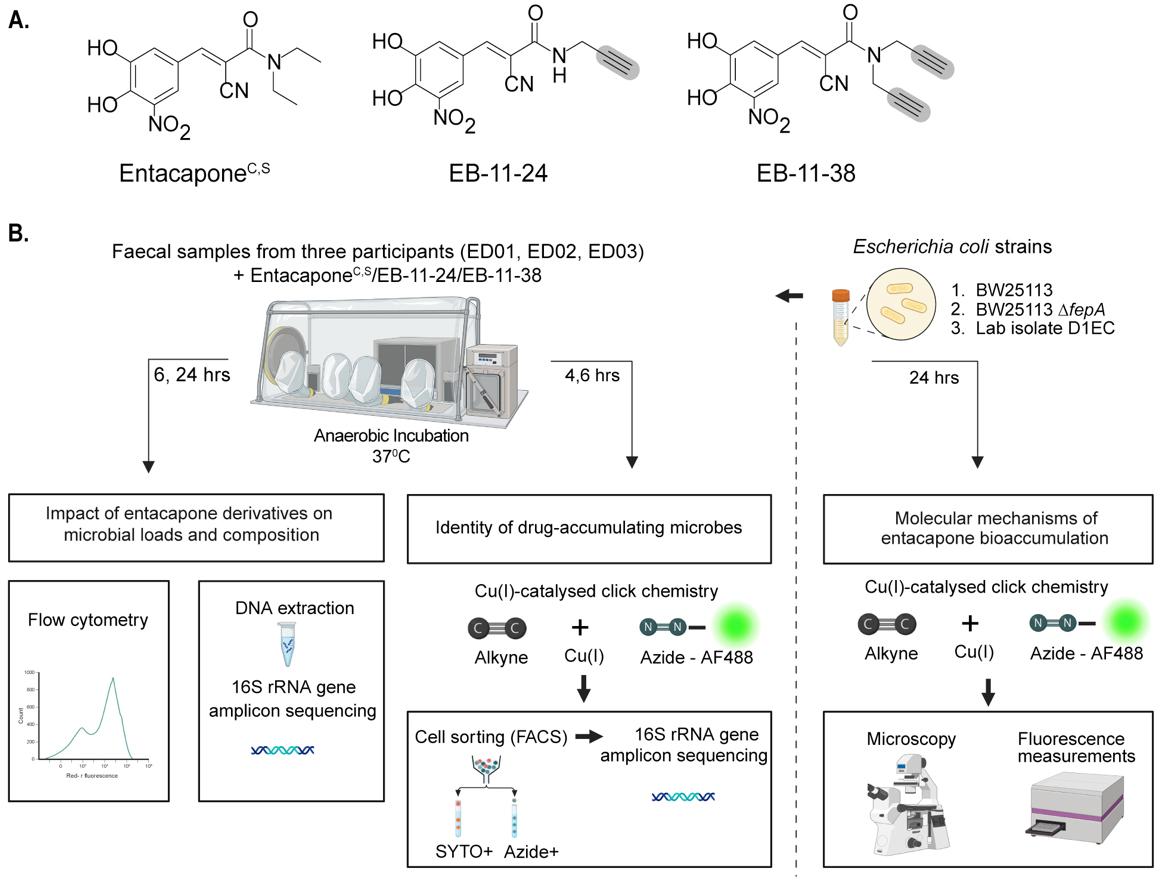
Figure 1. Bioorthogonal labelling combined with click chemistry to investigate entacapone bioaccumulation by microbes at the single-cell level. (A) Chemical structures of native entacapone (entacaponeC), and newly synthesised entacapone (entacaponeS) and alkynyl entacapone derivatives EB-11-24 (1 alkyne) and EB-11-38 (2 alkynes). Structural modifications to the native entacapone molecule include one or two terminal alkyne groups, highlighted in grey; (B) Ex vivo anaerobic incubation and processing of entacapone derivatives. Faecal samples from three healthy participants (ED01, ED02, ED03) were incubated under anaerobic conditions and supplemented with entacapone or its derivatives. Microbiome composition and abundance were assessed through flow cytometry and 16S rRNA gene sequencing at 6- and 24-h timepoints. Drug-responsive microbes were identified using click-chemistry and fluorescence-activated cell sorting (FACS) at 4- and 6-h timepoints, followed by 16S rRNA gene sequencing. E. coli strains were incubated with entacapone derivatives under anaerobic conditions and subjected to click-chemistry and fluorescence measurements to determine molecular cues of entacapone accumulation. This figure was created in Biorender (https://BioRender.com/xhx4f6v). rRNA: Ribosomal RNA; FACS: fluorescence-activated cell sorting.