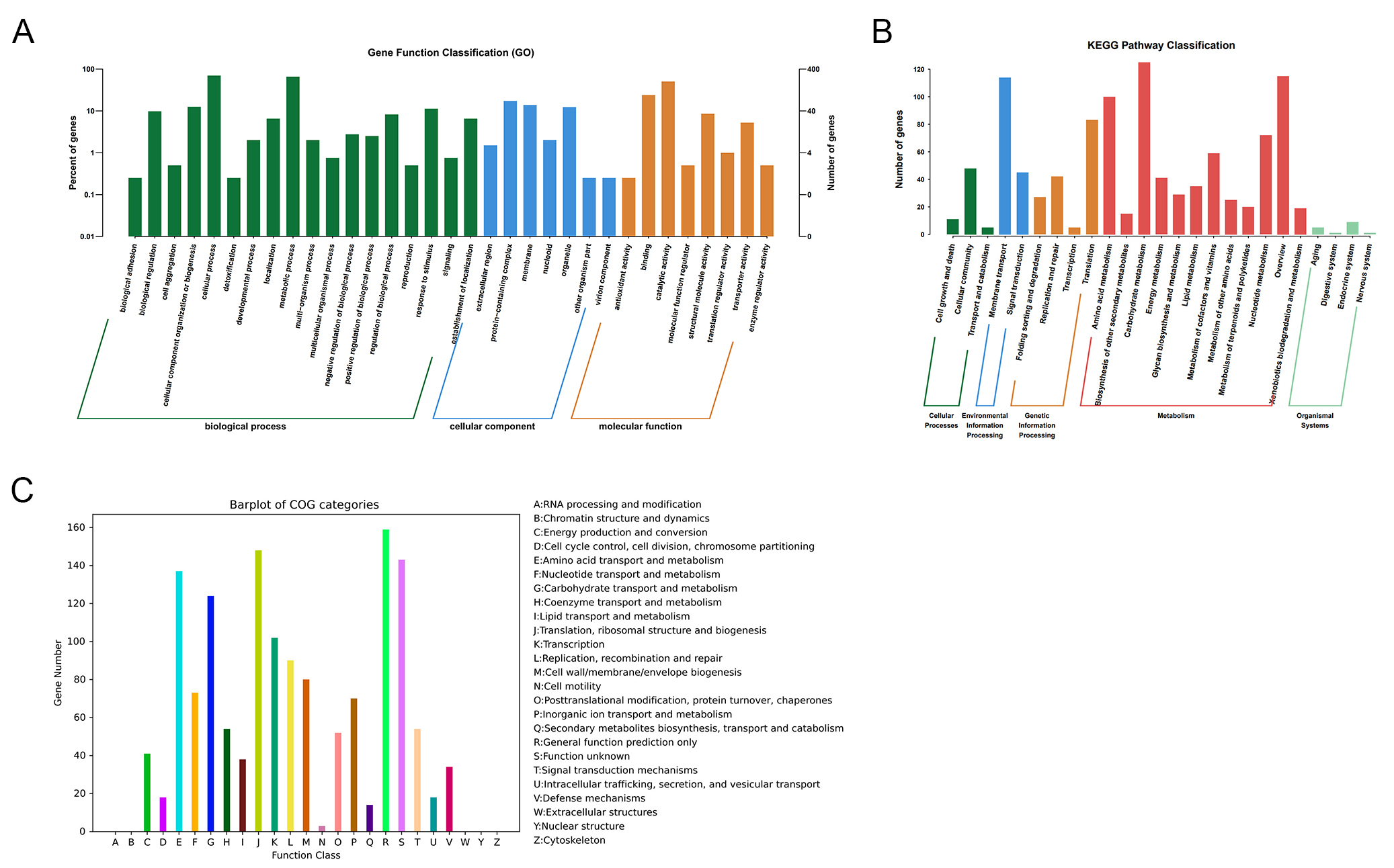
fig2
Figure 2. Functional annotation of the CRC211 genome. (A) GO classification of annotated genes across three main categories: Biological Process, Cellular Component, and Molecular Function; (B) KEGG pathway enrichment analysis showing the distribution of genes across metabolic pathways. The most enriched pathways were amino acid metabolism, carbohydrate metabolism, and overview; (C) COG functional categorization of predicted proteins. Major categories include: Translation, ribosomal structure, and biogenesis (J): protein synthesis; Amino acid transport and metabolism (E): nutrient uptake and utilization; Cell wall/membrane/envelope biogenesis (M): bacterial integrity and host interaction; Transcription (K), and Replication, recombination, and repair (L): genetic information processing. GO: Gene Ontology; COG: Clusters of Orthologous Groups.