fig7
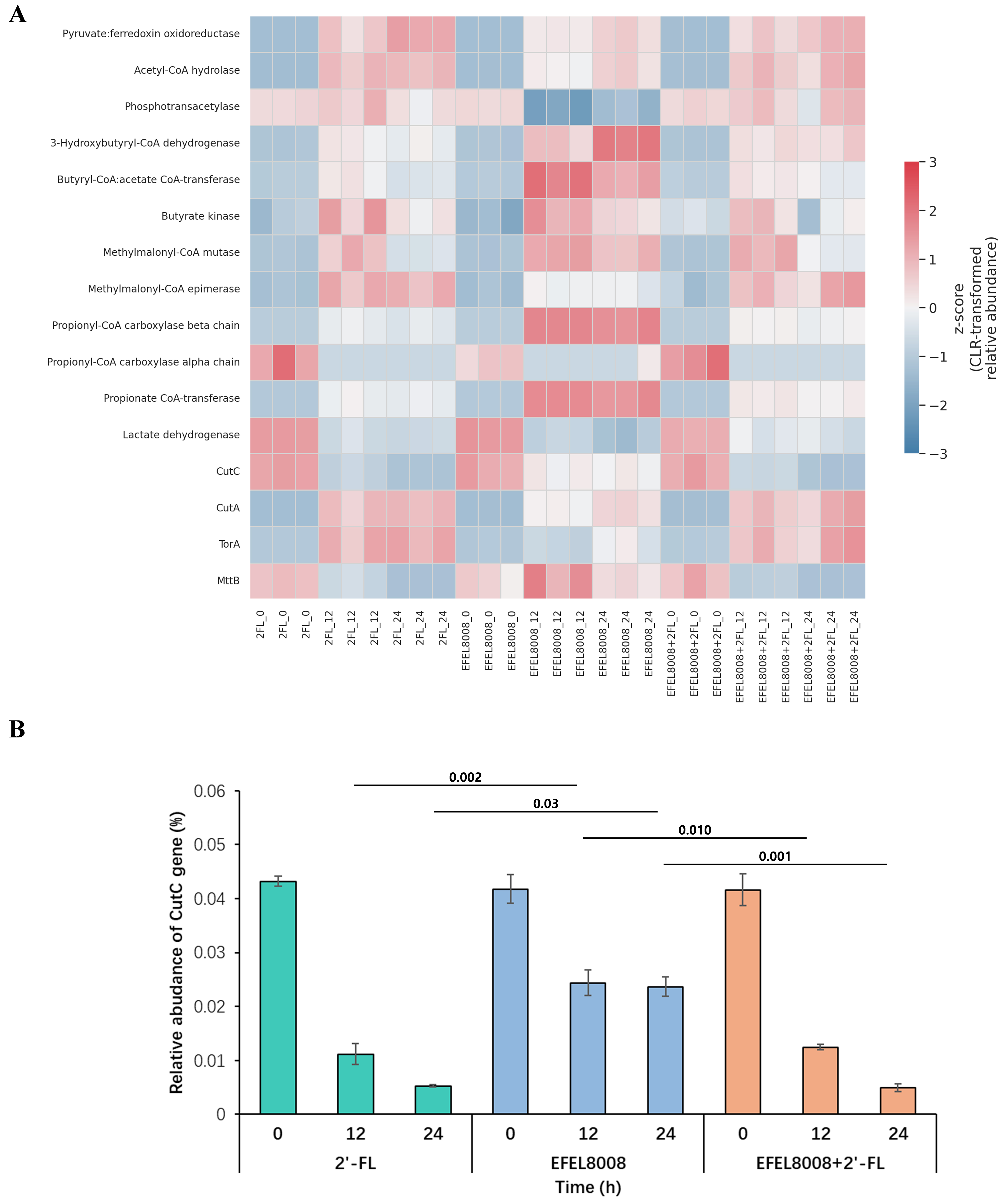
Figure 7. Functional prediction of microbial genes associated with SCFA metabolism and TMA production using PICRUSt2. (A) Heatmap showing the CLR-transformed Z-scores of KEGG orthologs related to microbial metabolic pathways during in vitro fecal fermentation. Genes involved in SCFA metabolism and TMA production are visualized across experimental groups and time points; (B) Relative abundance of the CutC gene across fermentation groups at 0, 12, and 24 h. Error bars indicate the standard deviation of three biological replicates (n = 3). Statistical significance was evaluated using t-test, and P-values are indicated above the bars. SCFA: Short-chain fatty acid; TMA: trimethylamine; CLR: centered log-ratio transformation; KEGG: Kyoto Encyclopedia of Genes and Genomes.