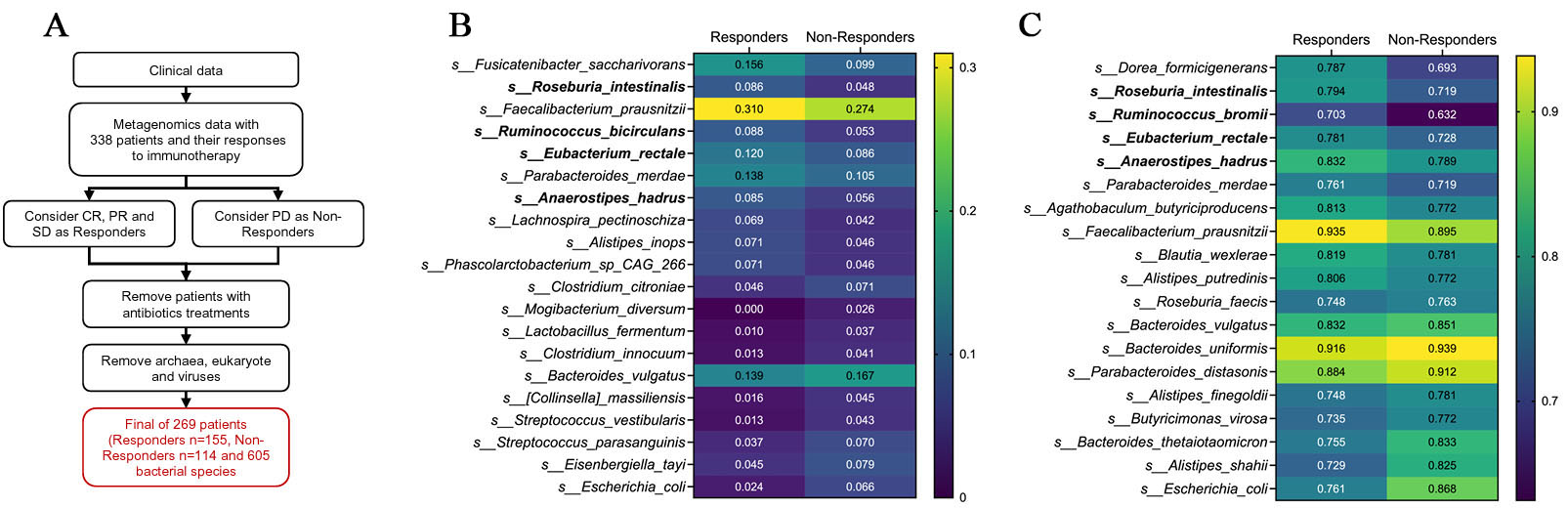
fig1
Figure 1. Abundance- and prevalence-based screening of response-associated bacterial species. (A) Schematic overview of the metagenomic re-analysis pipeline. Shotgun metagenomic data from patients with advanced NSCLC treated with anti-PD-1 therapy were re-analyzed. After exclusion of patients with recent antibiotic exposure and removal of non-bacterial features, 269 patients [responders (R), n = 155; non-responders (NR), n = 114] and 605 bacterial species were retained for downstream analysis; (B) Heatmap of mean relative abundance for species ranked by abundance mean difference (Δabundance = meanResponder - meanNon-responder) calculated from feature-wise min-max-scaled values. The top 10 responder-enriched and top 10 non-responder-enriched species are shown using the original relative abundance data; (C) Heatmap of prevalence for species ranked by prevalence difference (Δprevalence = prevResponder - prevNon-responder). Among species with prevalence ≥ 70% in responders, the top responder- and non-responder-enriched taxa are shown. CR: Complete response; PR: partial response; SD: stable disease; PD: progressive disease; NSCLC: non-small-cell lung cancer.