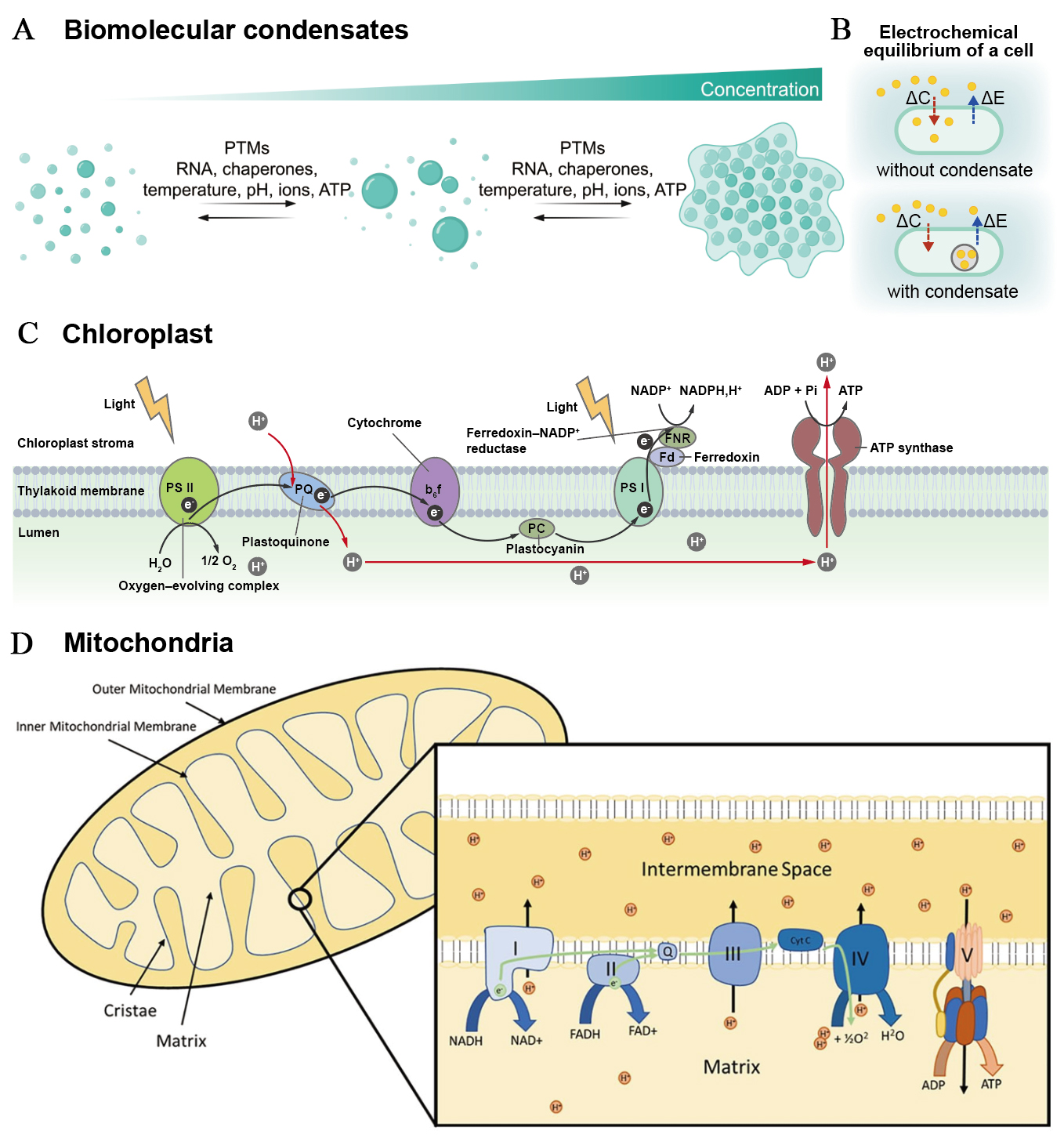
fig2
Figure 2. Molecular and subcellular ion dynamics. (A) Mechanisms of biomolecular condensation via phase separation: intrinsically disordered regions and RNA scaffolds form multivalent interactions to drive nucleation and subsequent protein-RNA recruitment. Post-translational modifications (PTMs) tune assembly/disassembly by altering interaction surfaces, while temperature, pH, ionic strength, and crowding conditions set phase-separation thresholds. This figure is quoted with permission from Choi et al.[37]; (B) Diagram showing how condensate formation shifts the intra- versus extracellular electrochemical balance, impacting membrane voltage; (C) Linear electron transport across the thylakoid: PSII uses light energy to oxidise water and reduce plastoquinone, cytochrome b6f transfers electrons to plastocyanin while releasing protons to the lumen, PSI reduces ferredoxin and FNR generates NADPH; the resulting proton motive force powers ATP synthase; (D) Overview of mitochondrial oxidative phosphorylation organisation: electron carriers feed complexes I-IV, protons are pumped across the inner membrane to establish a proton electrochemical gradient, and ATP synthase at cristae edges uses this gradient; carriers and supercomplexes coordinate flux and regulation. This figure is quoted with permission from Middleton et al.[38]. ΔC: Change in concentration; ΔE: change in electrochemical potential; NADP+: nicotinamide adenine dinucleotide phosphate; PSI: photosystem I; PSII: photosystem II; FNR: ferredoxin-NADP+ reductase; PC: plastocyanin; ADP: adenosine diphosphate; NADPH: reduced nicotinamide adenine dinucleotide phosphate; PQ: plastoquinone; NADH: reduced nicotinamide adenine dinucleotide; NAD: nicotinamide adenine dinucleotide; FADH: reduced flavin adenine dinucleotide; FAD: flavin adenine dinucleotide.