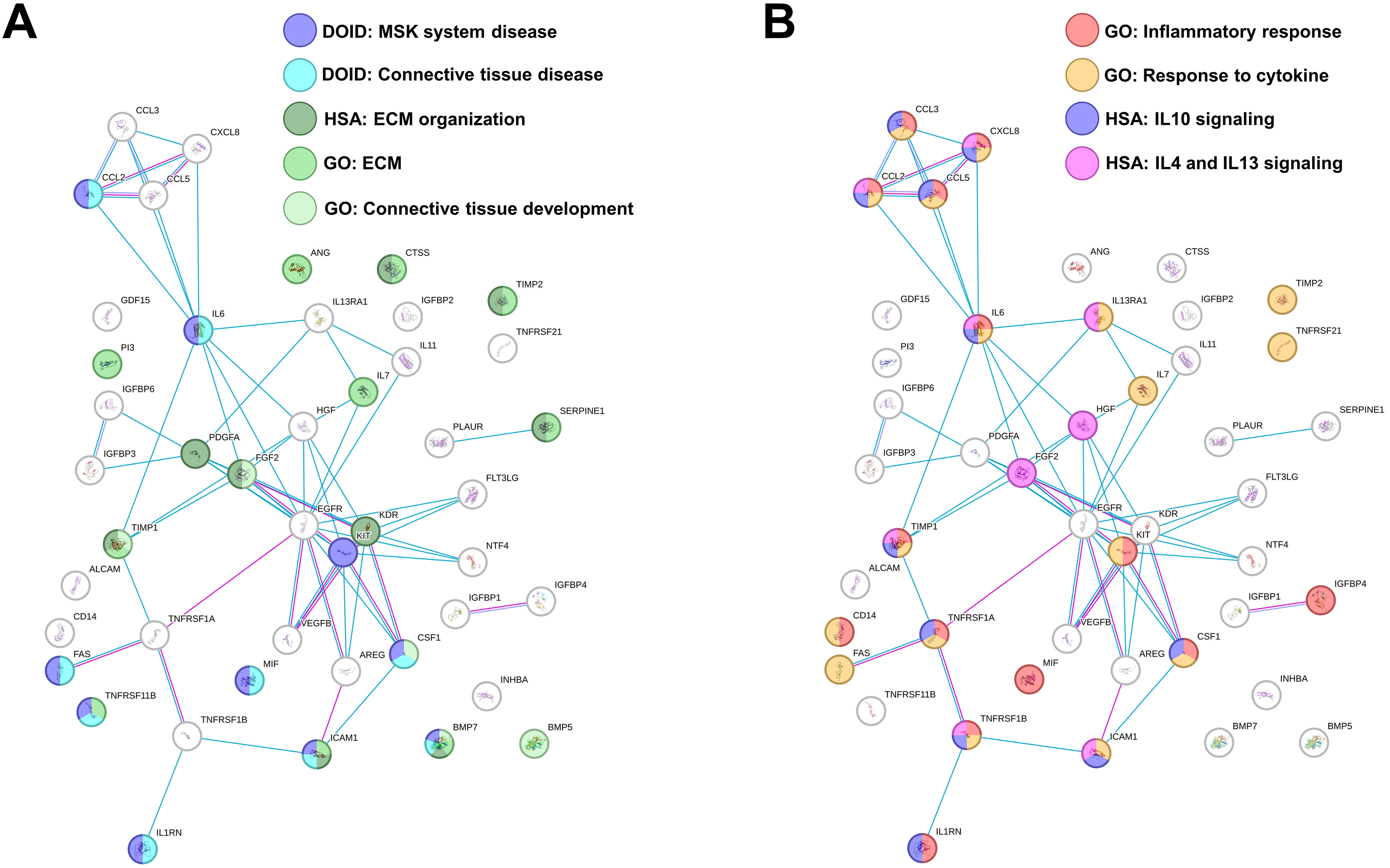
fig2
Figure 2. Functional association network for identified secreted factors. (A) Protein-protein interaction levels for proteins shared in MSC secretome, mined using STRING, and related to MSK and ECM terms. Blue connections: Proteins with known interactions based on curated databases; violet connections: proteins with experimentally determined interactions; Empty nodes: proteins of unknown 3D structure; filled nodes: known or predicted 3D structure; (B) Protein-protein interaction networks for proteins shared in MSC secretome, mined using STRING, and related to inflammation terms. For the color code, refer to (A). Data presented in PowerPoint. MSC: Mesenchymal stromal cell; MSK: musculoskeletal; ECM: extracellular matrix; GO: Gene Ontology; DOID: Disease Ontology ID; HSA: Homo sapiens pathway (Reactome); IL: interleukin; STRING: Search Tool for the Retrieval of Interacting Genes/Proteins.