fig3
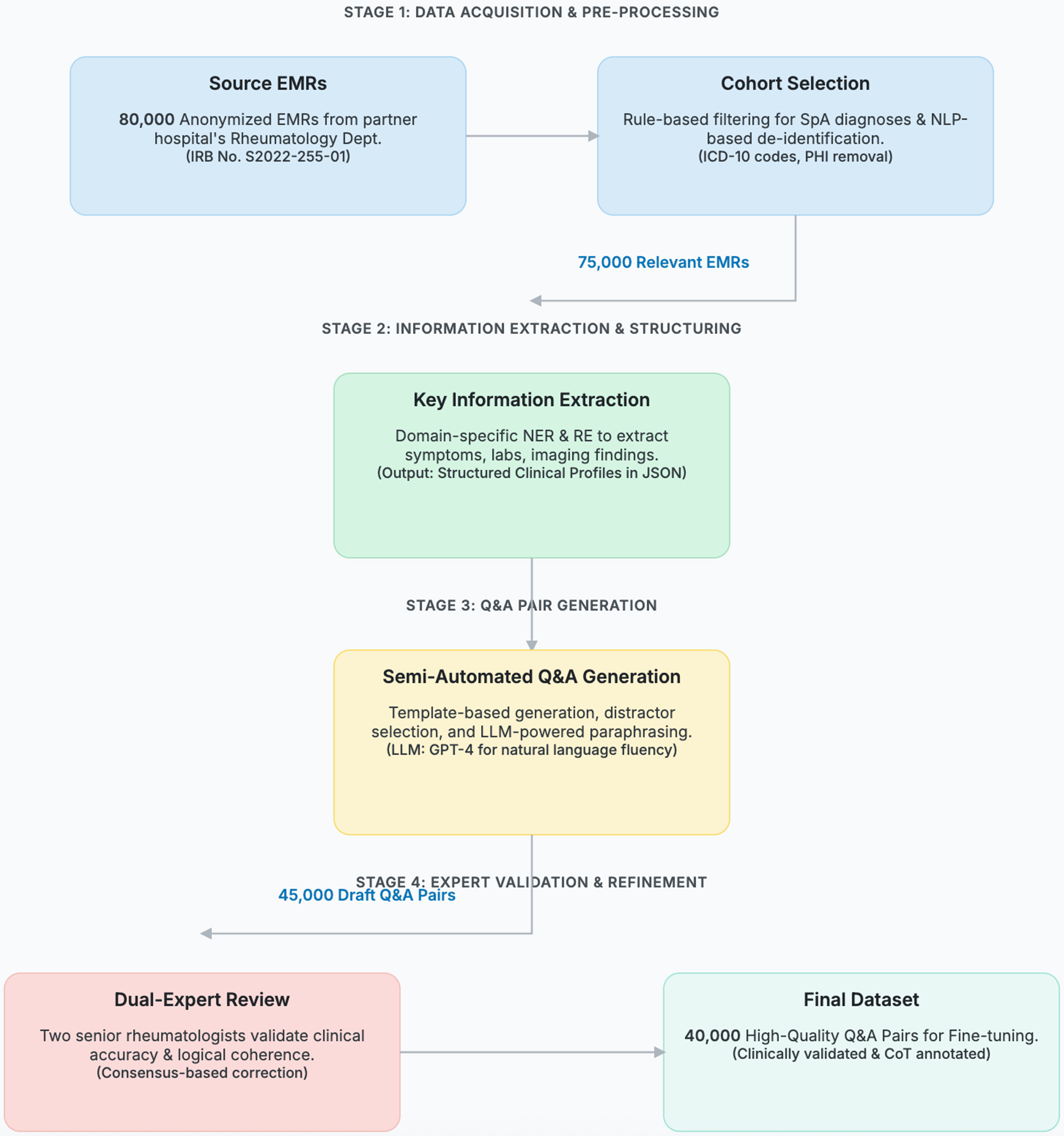
Figure 3. The end-to-end data curation workflow for transforming raw electronic medical records (EMRs) into a high-quality, fine- tuning dataset. The process is organized into four sequential stages: (1) Acquisition & Pre-processing, where 80,000 anonymized EMRs are selected and de-identified; (2) Information Extraction, where key clinical information is structured using domain-specific NLP models; (3) Q&A Generation, where draft question-answer pairs are semi-automatically created; and (4) Expert Validation, where two senior rheumatologists review, correct, and annotate each pair to ensure clinical accuracy and logical coherence. The numbers on the connecting arrows indicate the data volume after each key filtering step, culminating in the final 40,000-pair dataset. NLP: Natural language processing; IRB: Institutional Review Board; SpA: spondyloarthritis; ICD: International Classification of Diseases; PHI: Protected Health Information; NER: named entity recognition; RE: relation extraction; JSON: JavaScript Object Notation; LLM: Large Language Model; GPT-4: Generative Pre-trained Transformer 4; CoT: Chain-of-Thought.