fig5
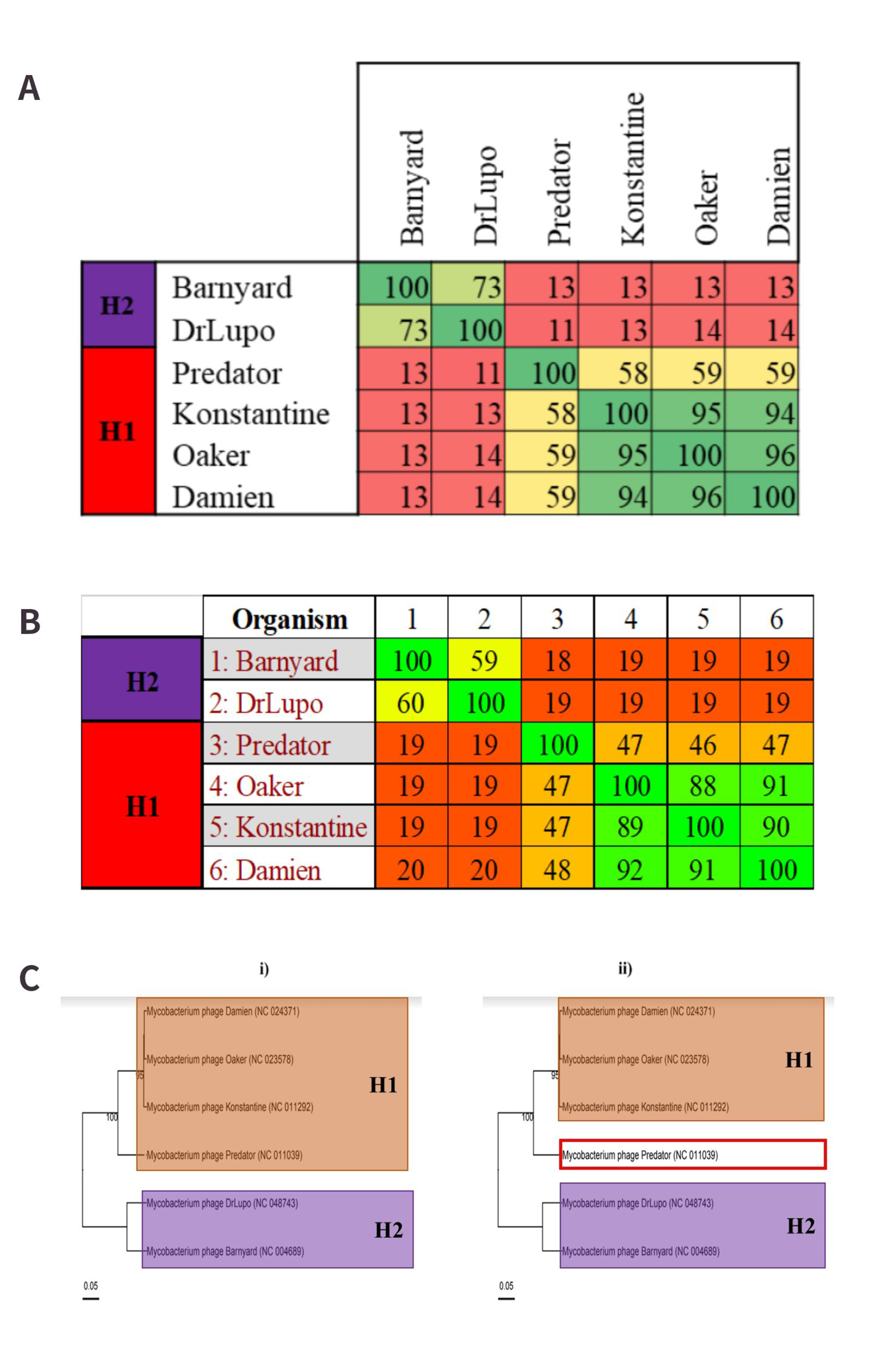
Figure 5. Proposed cluster assignments for cluster H. (A) VIRIDIC alignment of cluster H. The single Predatorvirus, Predator, does not meet the ≥ 70% nucleotide similarity threshold for inclusion with the larger H1 genus, Konstantinevirus; (B) Gegenees proteome alignment for cluster H. Predator also does not meet the proposed minimum proteome similarity threshold of 50% for inclusion in the same subcluster as any member of the Konstantineviruses; (C) (i) Existing subcluster classifications of cluster H. H1 and H2 are observable on two very distinct clades. Predator shares a clade with the other H1 viruses. (ii) Proposed subcluster creation for Predatorviruses. While Predator shares a clade with the Konstantineviruses, the divergence of its branch from the other phages would appear to reflect the < 50% proteome similarity noted in the Gegenees output, which would support the creation of a novel subcluster (indicated in the red square).









