Association of PTPRC gene polymorphisms with TNF inhibitor efficacy in rheumatoid arthritis: a systematic review and meta-analysis
Abstract
Background: Rheumatoid arthritis (RA) is a chronic autoimmune inflammatory disease characterized by joint destruction and functional impairment. Tumor Necrosis Factor inhibitors (TNFi) are widely used biologic therapies for RA; however, patient outcomes show significant individual variation. Genetic factors, including polymorphisms in the PTPRC gene, have been investigated as potential predictors of TNFi efficacy, but results remain inconsistent.
Methods: We conducted a systematic review and meta-analysis following PRISMA guidelines to evaluate the association between PTPRC polymorphisms and TNFi treatment response in RA patients. A comprehensive search was performed in PubMed and Scopus databases up to April 2025. Studies were screened and quality assessed using the Q-Genie tool. Meta-analyses employed Mantel-Haenszel methods with fixed and random effects models.
Results: Five studies met eligibility criteria, including data from 12 European cohorts totaling 1,543 RA patients. The main polymorphism assessed was rs10919563. Meta-analysis revealed the rs10919563 A allele was significantly associated with increased odds of non-response to TNFi (allelic model OR: 1.94; 95%CI: 1.31-2.88). The recessive models supported these findings, and no publication bias was detected.
Conclusions: Our results consolidate evidence supporting the PTPRC rs10919563 polymorphism as a potential predictive genetic biomarker for TNFi efficacy in RA, which could be integrated into personalized treatment strategies. Further studies are required to validate clinical utility across diverse populations.
Keywords
INTRODUCTION
Rheumatoid arthritis (RA) is a chronic autoimmune inflammatory disease characterized by progressive joint destruction, leading to pain, stiffness, and significant functional impairment[1]. RA is estimated to affect approximately 0.5% to 1% of the global population, with a considerable impact on patients' quality of life and substantial socioeconomic costs associated with its management[1,2]. Tumor Necrosis Factor inhibitors (TNFi) represent one of the main classes of biologic therapies used in the treatment of RA, providing symptom relief and slowing disease progression[3,4]. However, the response to TNFi varies significantly among patients, with a substantial proportion exhibiting only a partial response or no response to treatment[5]. This variability has driven the search for genetic biomarkers capable of predicting TNFi efficacy and enabling a more personalized approach to RA management[6-9].
The PTPRC gene (Protein Tyrosine Phosphatase Receptor Type C), also known as CD45, encodes a protein that is essential for regulating lymphocyte activation and plays a fundamental role in the adaptive immune response[10,11]. Genetic polymorphisms in this gene have been implicated in the modulation of immune activity and may influence the response to biologic therapies, including Tumor Necrosis Factor inhibitors[7,12,13]. Functionally, PTPRC encodes the CD45 phosphatase, which regulates T and B cell receptor signaling. Polymorphisms such as rs10919563 may alter CD45 expression or activity, potentially modulating downstream TNF signaling pathways and influencing patient response to TNF inhibitor therapy[14].
Several studies have investigated the association between the PTPRC rs10919563 polymorphism and the efficacy of TNFi treatment in rheumatoid arthritis, but the findings remain inconsistent. While some studies found a significant association between the G allele and improved TNFi response[15,16], others reported no association[17,18]. In a multicenter cohort study, heterogeneity in results among different populations was also reported[19]. These discrepancies may be attributed to differences in study design, sample size, ethnicity of the populations evaluated, clinical response criteria, and the type of TNF inhibitor used, highlighting the need for a more robust and updated systematic analysis. It should be noted that all available cohorts included in these studies were composed predominantly of European or European descendant populations. This geographic homogeneity limits the generalizability of findings to non-European populations, which constitute the majority of global RA patients. Differences in genetic diversity across populations may influence expected effect sizes and treatment response, highlighting the need for caution when extrapolating these results to other ethnic groups.
Two meta-analyses published in 2016 and 2017 evaluated the association between genetic variants, including the PTPRC gene, and treatment response to anti-TNF therapy in patients with rheumatoid arthritis[12,20]. The 2016 meta-analysis found a significant association between the A allele of the rs10919563 polymorphism in PTPRC and a reduced likelihood of response to TNF-α inhibitor treatment, suggesting its potential as a predictive biomarker[20]. The 2017 meta-analysis, which reviewed a broader set of genetic variants associated with anti-TNF response, confirmed the involvement of PTPRC among other genes related to inflammatory signaling and lymphocyte activation, although it emphasized that robust clinical biomarkers for therapeutic selection were not yet available[12].
In light of this, we conducted a systematic review and meta-analysis to assess the association between PTPRC gene polymorphisms and the response to TNF inhibitors in patients with rheumatoid arthritis. Our objective is to consolidate the available evidence and update the existing knowledge since the last published reviews - particularly the 2017 meta-analysis[20] - providing a more comprehensive and current overview of the potential role of PTPRC as a predictive genetic biomarker for TNFi efficacy in RA.
METHODS
This systematic review was registered on the PROSPERO platform (CRD420251079455), following international guidelines for systematic reviews. Searches were conducted in the PubMed and Scopus databases, covering relevant studies published up to April 30, 2025. The selection and analysis process adhered to the PRISMA guidelines (Preferred Reporting Items for Systematic Reviews and Meta-Analyses), ensuring methodological transparency and reproducibility of the findings.
The search strategy employed Boolean logic, combining terms related to: (i) rheumatoid arthritis
Studies were included if they met the following criteria: (i) publication in English; (ii) investigation of genetic associations involving the PTPRC gene and therapeutic response to anti-TNF drugs in rheumatoid arthritis. Genome-wide association studies (GWAS), systematic reviews, meta-analyses, and studies not addressing the genetics of rheumatoid arthritis were excluded. Study screening was independently conducted by two reviewers, and disagreements were resolved by consensus.
Quality assessment of the studies
The methodological quality of the included studies was evaluated by two independent reviewers using the Q-Genie tool[21-23], which considers criteria such as the scientific justification of the research, definition of comparison groups, sample size, and description of planned statistical analyses. Since all studies had a case-control design, the quality classification followed previously proposed criteria (scores ≤ 35 indicate low quality, > 35 and ≤ 45 moderate quality, and > 45 good quality, on a scale ranging from 7 to 77 points)[21].
Statistical analyses
Statistical analyses and all main figures were performed/generated using R software (version 4.2.1). Hardy-Weinberg equilibrium was assessed in both case and control groups for each cohort included in the meta-analysis using Fisher’s exact test. The combined evaluation of polymorphisms was conducted through meta-analysis using the Mantel-Haenszel method, applying fixed- and random-effects models via the metabin function[24]. A significance level of 5% was adopted. To assess publication bias, Egger’s test and visual inspection of the funnel plot were used.
RESULTS
A total of five studies evaluating the association between PTPRC gene polymorphisms and therapeutic response to tumor necrosis factor inhibitors (TNFi) in patients with rheumatoid arthritis were deemed eligible after the screening process. Of these, four studies were included in the meta-analysis, encompassing 12 distinct cohorts. All cohorts were from European populations, including Spain, Greece, the Netherlands, the United Kingdom, Sweden, and Portugal, as well as individuals of European descent from the USA [Figure 1]. Quality assessment using the Q-Genie tool classified all five studies as good quality, with a mean score of 50.46 and a standard deviation of 3.82, indicating overall good methodological quality. The item with the lowest average rating was item 3, "Selection and comparability of comparison groups”, with a mean score of 2.7 (SD = 1.1), suggesting potential limitations in group matching or representativeness. In contrast, the best-rated items were item 1, "Rationale for study”, and item 11, "Appropriateness of inferences drawn from results”, with mean scores of 6.36 (SD = 0.40) and 6.42 (SD = 0.58), respectively, indicating strong conceptual justification and appropriate interpretation of findings [Supplementary Table 1]. The results of the Hardy-Weinberg equilibrium tests for the cohorts included in the meta-analysis are presented in Supplementary Table 2.
Figure 1. PRISMA flow diagram illustrating the study selection process for the systematic review and meta-analysis. Original figure created by the authors for this study.
It is noteworthy that one study was excluded from the meta-analysis because it included patients from the BRAGGSS cohort, which overlapped with another included cohort[16,19]. This exclusion was made to avoid sample overlap and ensure independence of the data analyzed. A minimum of three studies was required to perform each meta-analysis.
The anti-TNF agents used in the included studies were etanercept (ETAN), infliximab (INFLIX), adalimumab (ADALI), and golimumab (GOLI), with only the study by Canhão et al. (2015) including patients treated with golimumab[18]. All studies employed the Disease Activity Score 28 (DAS28) as the criterion for therapeutic response assessment, with a DAS28 score of ≤ 3.2 considered indicative of a good response. Furthermore, to ensure comparability, only studies evaluating response after a minimum period of six months were included.
Two polymorphisms (rs10919563 and rs6683595) in the PTPRC gene were reported. However, only one study assessed the association between the rs6683595 polymorphism and response to anti-TNF inhibitors[15].
Thus, a meta-analysis was conducted only for the rs10919563 polymorphism and non-response to anti-TNF therapy, with response evaluated using the DAS28. Effect sizes from 12 distinct cohorts were combined, nine of which were reported in one study[19] [Table 1]. A total of 552 non-responders and 991 responders were included. Funnel plot asymmetry was visually assessed for all three genetic models. Egger’s test indicated no evidence of publication bias for the allelic model (t = 0.33, P = 0.746), the recessive allele A model (t = -0.73, P = 0.485), or the recessive allele G model (t = -1.29, P = 0.225) [Supplementary Figure 1].
Characteristics of the included studies that evaluated the association between TNF inhibitors and the rs10919563 or rs6683595 polymorphisms
| Autor | Year | Country | Sample | Polymorphism | Anti-TNF agent | RAT* | RAC* | Q-Genie assessment |
| Cui (ABCoN) | 2010 | USA | 75 | rs10919563 | ETAN/INFLIX/ADALI | 6 months | DAS28 | Good |
| Cui (AMC) | 2010 | Netherlands | 79 | rs10919563 | INFLIX/ADALI | 6 months | DAS28 | Good |
| Cui (BeSt) | 2010 | USA | 126 | rs10919563 | INFLIX | 6 months | DAS28 | Good |
| Cui (BRAGGSS) | 2010 | UK | 74 | rs10919563 | INFLIX | 6 months | DAS28 | Good |
| Cui (BRASS) | 2010 | USA | 44 | rs10919563 | ETAN/INFLIX/ADALI | 6 months | DAS28 | Good |
| Cui (EIRA) | 2010 | Swedish | 186 | rs10919563 | ETAN/INFLIX/ADALI | 6 months | DAS28 | Good |
| Cui (eRA) | 2010 | USA | 116 | rs10919563 | ETAN | 6 months | DAS28 | Good |
| Cui (KI) | 2010 | Swedish | 74 | rs10919563 | ETAN/INFLIX/ADALI | 6 months | DAS28 | Good |
| Cui (JBI) | 2010 | Netherlands | 43 | rs10919563 | ETAN | 6 months | DAS28 | Good |
| Plant | 2012 | UK | 469 | rs10919563 | ETAN/INFLIX/ADALI | 6 months | DAS28 | Good |
| Zervou | 2013 | Greece | 193 | rs10919563 | ETAN/INFLIX/ADALI | 6 months | DAS28 | Good |
| Ferreiro-Iglesias | 2015 | Spain | 398 | rs10919563 and rs6683595 | ETAN/INFLIX/ADALI | 6 months | DAS28 | Good |
| Canhão | 2015 | Portugal | 135 | rs10919563 | ETAN/INFLIX/ADALI/GOLI | 6 months | DAS28 | Good |
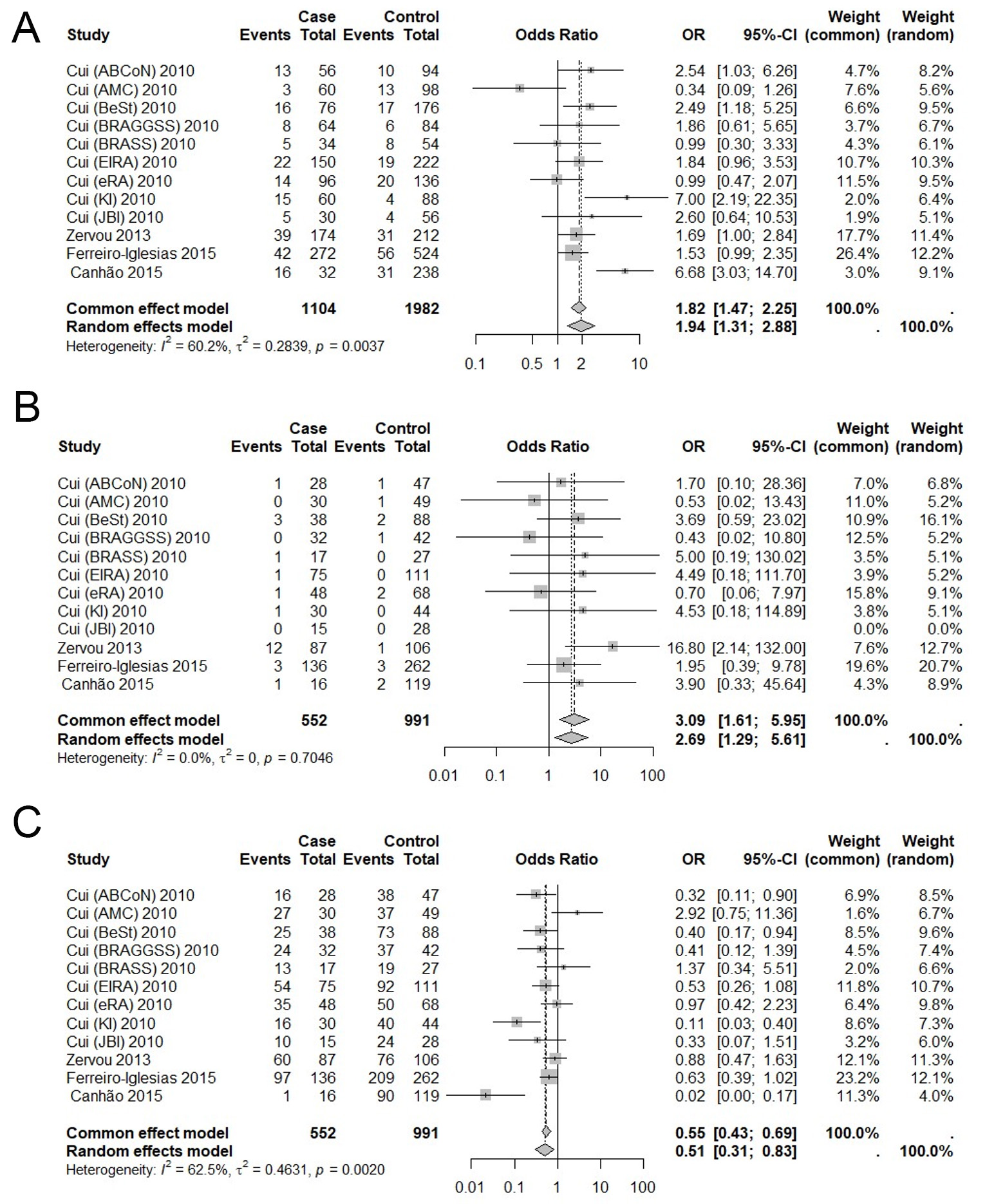
The three genetic models showed statistically significant associations: the allelic and recessive allele G models were analyzed using a random-effects model, while the recessive allele A model was analyzed using a fixed-effects model. The allelic model (risk allele A) was associated with a 94% increased odds of treatment non-response (OR: 1.94; 95%CI: 1.31-2.88) [Figure 2A]. The recessive model for allele A also showed increased odds of non-response (OR: 3.09; 95%CI: 1.61-5.95) [Figure 2B]. Conversely, the recessive model for allele G was associated with a 49% reduction in the odds of non-response (OR: 0.51; 95%CI: 0.31-0.83) [Figure 2C].
Figure 2. Meta-analysis of the association between the rs10919563 polymorphism in the PTPRC gene and response to TNF inhibitor treatment in patients with rheumatoid arthritis. The three genetic models showed statistically significant associations: the allelic model (A), the recessive model for allele A (B), and the recessive model for allele G (C). The allelic and recessive models of the G allele were analyzed using a random-effects model, while the recessive model was analyzed using a fixed-effects model. The combined analysis of the polymorphisms was performed using the Mantel-Haenszel meta-analysis, applying fixed- and random-effects models through the metabin function. Original figure created by the authors for this study.
DISCUSSION
This meta-analysis updated the available evidence on the association between the rs10919563 polymorphism in the PTPRC gene and therapeutic response to Tumor Necrosis Factor inhibitors (TNFi) in patients with rheumatoid arthritis (RA). The results showed a significant association between the A allele and an increased likelihood of non-response to treatment, with consistency across all three genetic models analyzed and no evidence of publication bias. These findings reinforce the potential of PTPRC as a predictive genetic biomarker of TNFi efficacy, which may contribute to more personalized strategies in RA management.
Among the included studies, only one evaluated, in addition to the rs10919563 polymorphism, the rs6683595 polymorphism, which is also located in an intronic region of the PTPRC gene[15]. However, no significant association was observed between this second polymorphism and the response to TNFi treatment[15]. Considering the potential regulatory impact of intronic variants on gene expression, it would be relevant for future studies to attempt to replicate the analysis of rs6683595 in other independent cohorts to clarify its role in the context of rheumatoid arthritis pharmacogenetics[25-27].
The PTPRC gene encodes the CD45 protein, a membrane-associated tyrosine phosphatase widely expressed in hematopoietic cells, which is essential for T and B lymphocyte activation through modulation of antigen receptor signaling[28-30]. Polymorphisms such as rs10919563 may influence the expression or function of CD45, thereby altering the activation threshold of these immune cells[7]. Given that TNF inhibitors act on the inflammatory cascade that follows lymphocyte activation, it is plausible that variants in PTPRC may indirectly affect TNFi efficacy by modulating baseline immune activation or the persistence of the inflammatory response[2,3,31,32]. Thus, the association observed in this meta-analysis is supported by known immunological mechanisms, which reinforces the relevance of PTPRC as a potential predictive genetic biomarker in rheumatoid arthritis. Ideally, the meta-analysis would be stratified by specific TNF inhibitors, since - although all classified as TNFi - these agents differ in molecular structure and mechanisms of action, which may influence therapeutic response[33]. However, the included studies did not provide stratified outcome data for each individual drug; instead, they grouped all therapies under the general category of "TNF inhibitors”, only listing the agents used. This lack of stratification limits the ability to determine whether the observed associations with PTPRC variants are driven by a particular agent or are consistent across all TNFi.
Two previous meta-analyses, published in 2016 and 2017, had already explored the relationship between genetic variants and the response to TNFi, including the PTPRC gene[12,20]. However, both included a study that was excluded from the present analysis due to sample overlap with another cohort[16,19]. This exclusion was essential to avoid data duplication and ensure the statistical independence of the analyzed samples, even though it reduced the total number of studies included. Although no new publications on PTPRC and the response to TNFi have been identified since the last meta-analysis in 2017, this may reflect a shift in research focus toward other genetic markers or alternative therapeutic modalities. In addition, studies involving non-European populations or reporting null associations may remain unpublished due to publication bias, which could explain the predominance of European cohorts in the literature. The present review updates and consolidates existing knowledge based on more rigorous methodological criteria. In addition, it applied updated tools for assessing study quality (Q-Genie) and controlling for publication bias, which enhances the robustness of the evidence presented[21,23].
An important limitation to highlight is the geographic restriction of the included cohorts, all of which were derived from European populations. This limits the generalizability of the findings to other populations, such as Latin Americans, Africans, or Asians[34,35]. Although one cohort included 57 individuals with self-reported Greek Caucasian ancestry, all patients were recruited and treated in hospitals in Spain[15]. Considering the low proportion of these individuals and the absence of stratified ancestry data, the sample was regarded as a single population predominantly of Spanish origin. Furthermore, population stratification was controlled in the original study through ancestry covariates, which reduces the likelihood of bias[15].
The absence of new publications since 2017 addressing the relationship between PTPRC and response to TNFi may reflect a significant gap in the literature. Future studies are needed to validate these findings in more heterogeneous cohorts and across different clinical and ethnic contexts. Additionally, functional investigations elucidating the biological mechanisms related to the rs10919563 polymorphism could strengthen its applicability as a biomarker. Integrating genetic information with clinical data may represent a significant step forward toward personalized medicine in rheumatoid arthritis.
DECLARATIONS
Acknowledgments
The authors would like to thank Canva (www.canva.com) for generating the graphical abstract, which aided in visualizing the main findings of this study.
Authors’ contributions
Conducted data analysis, study screening, quality assessment, drafted the first version of the manuscript, and contributed to subsequent revisions: de Araújo JLF
Assisted in study screening, quality assessment, and contributed to the final manuscript revision: de Almeida IM
Contributed to quality assessment and reviewed the final manuscript: Azevedo SB, Landeiro LdSG
Performed the final review of the manuscript: Costa RdS
Availability of data and materials
All data generated or analyzed during this study are included in this published article and its
Financial support and sponsorship
None.
Conflict of interest
All authors declared that there are no conflicts of interest.
Ethical approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Copyright
© The Author(s) 2025.
Supplementary Materials
REFERENCES
2. Lin YJ, Anzaghe M, Schülke S. Update on the pathomechanism, diagnosis, and treatment options for rheumatoid arthritis. Cells. 2020;9:880.
3. Caporali R, Kadakia A, Howell O, et al. A real-world comparison of clinical effectiveness in patients with rheumatoid arthritis treated with upadacitinib, tumor necrosis factor inhibitors, and other advanced therapies after switching from an initial tumor necrosis factor inhibitor. Adv Ther. 2024;41:3706-21.
4. Mohammadi P, Hesari M, Chalabi M, Salari F, Khademi F. An overview of immune checkpoint therapy in autoimmune diseases. Int Immunopharmacol. 2022;107:108647.
5. Akiyama M, Kaneko Y. Pathogenesis, clinical features, and treatment strategy for rheumatoid arthritis-associated interstitial lung disease. Autoimmun Rev. 2022;21:103056.
6. Scott IC, Rijsdijk F, Walker J, et al. Do genetic susceptibility variants associate with disease severity in early active rheumatoid arthritis? J Rheumatol. 2015;42:1131-40.
7. Raychaudhuri S, Thomson BP, Remmers EF, et al. Genetic variants at CD28, PRDM1 and CD2/CD58 are associated with rheumatoid arthritis risk. Nat Genet. 2009;41:1313-8.
8. Barton A, Thomson W, Ke X, et al. Re-evaluation of putative rheumatoid arthritis susceptibility genes in the post-genome wide association study era and hypothesis of a key pathway underlying susceptibility. Hum Mol Genet. 2008;17:2274-9.
9. Orozco G, Martín J. Identification of new susceptibility markers for rheumatoid arthritis and systemic lupus erythematosus in the STAT4 gene. Per Med. 2008;5:169-74.
10. Mohren L, Doege A, Miroschnikov N, Dräger O, Busch MA, Dünker N. Role of protein tyrosine phosphatase receptor type E (PTPRE) in chemoresistant retinoblastoma. Int J Mol Sci. 2024;25:4572.
11. Lippert AH, Paluch C, Gaglioni M, et al. Antibody agonists trigger immune receptor signaling through local exclusion of receptor-type protein tyrosine phosphatases. Immunity. 2024;57:256-70.e10.
12. Bek S, Bojesen AB, Nielsen JV, et al. Systematic review and meta-analysis: pharmacogenetics of anti-TNF treatment response in rheumatoid arthritis. Pharmacogenomics J. 2017;17:403-11.
13. Prasad P, Kumar A, Gupta R, Juyal RC, Thelma BK. Caucasian and Asian specific rheumatoid arthritis risk loci reveal limited replication and apparent allelic heterogeneity in north Indians. PLoS One. 2012;7:e31584.
14. Raychaudhuri S, Remmers EF, Lee AT, et al. Common variants at CD40 and other loci confer risk of rheumatoid arthritis. Nat Genet. 2008;40:1216-23.
15. Ferreiro-Iglesias A, Montes A, Perez-Pampin E, et al. Replication of PTPRC as genetic biomarker of response to TNF inhibitors in patients with rheumatoid arthritis. Pharmacogenomics J. 2016;16:137-40.
16. Plant D, Prajapati R, Hyrich KL, et al. Replication of association of the PTPRC gene with response to anti-tumor necrosis factor therapy in a large UK cohort. Arthritis Rheum. 2012;64:665-70.
17. Zervou MI, Myrthianou E, Flouri I, et al. Lack of association of variants previously associated with anti-TNF medication response in rheumatoid arthritis patients: results from a homogeneous Greek population. PLoS One. 2013;8:e74375.
18. Canhão H, Rodrigues AM, Santos MJ, et al. TRAF1/C5 but not PTPRC variants are potential predictors of rheumatoid arthritis response to anti-tumor necrosis factor therapy. Biomed Res Int. 2015;2015:490295.
19. Cui J, Saevarsdottir S, Thomson B, et al. Rheumatoid arthritis risk allele PTPRC is also associated with response to anti-tumor necrosis factor alpha therapy. Arthritis Rheum. 2010;62:1849-61.
20. Lee YH, Bae SC. Associations between PTPRC rs10919563 A/G and FCGR2A R131H polymorphisms and responsiveness to TNF blockers in rheumatoid arthritis: a meta-analysis. Rheumatol Int. 2016;36:837-44.
21. Sohani ZN, Anand S, Meyre D, et al. Q-Genie. 2015; pp. 1-4. Available from: http://fhs.mcmaster.ca/pgp/links.html [Last accessed on 29 Sep 2025].
22. Sohani ZN, Sarma S, Alyass A, et al. Empirical evaluation of the Q-Genie tool: a protocol for assessment of effectiveness. BMJ Open. 2016;6:e010403.
23. Sohani ZN, Meyre D, de Souza RJ, et al. Assessing the quality of published genetic association studies in meta-analyses: the quality of genetic studies (Q-Genie) tool. BMC Genet. 2015;16:50.
24. Schwarzer G, Carpenter JR, Rücker G. Meta-analysis with R. Cham: Springer International Publishing; 2015.
26. Vaz-Drago R, Custódio N, Carmo-Fonseca M. Deep intronic mutations and human disease. Hum Genet. 2017;136:1093-111.
27. Ingelman-Sundberg M, Nebert DW, Lauschke VM. Emerging trends in pharmacogenomics: from common variant associations toward comprehensive genomic profiling. Hum Genomics. 2023;17:105.
28. Whyte ML, Smith KA, Buchberger A, et al. The roseoloviruses downregulate the protein tyrosine phosphatase PTPRC (CD45). J Virol. 2021;95:e0162820.
29. Alborzian Deh Sheikh A, Akatsu C, Abdu-Allah HHM, et al. The protein tyrosine phosphatase SHP-1 (PTPN6) but not CD45 (PTPRC) is essential for the ligand-mediated regulation of CD22 in BCR-ligated B cells. J Immunol. 2021;206:2544-51.
30. Barashdi MA, Ali A, McMullin MF, Mills K. Protein tyrosine phosphatase receptor type C (PTPRC or CD45). J Clin Pathol. 2021;74:548-52.
31. Aletaha D, Smolen JS. Diagnosis and management of rheumatoid arthritis: a review. JAMA. 2018;320:1360-72.
32. Stepanov AA, Malsagova KA, Kopylov AT, et al. Determination of heterogeneous proteomic and metabolomic response in anti-TNF and anti-IL-6 treatment of patients with rheumatoid arthritis. Life. 2023;13:596.
33. Furst DE, Wallis R, Broder M, Beenhouwer DO. Tumor necrosis factor antagonists: different kinetics and/or mechanisms of action may explain differences in the risk for developing granulomatous infection. Semin Arthritis Rheum. 2006;36:159-67.
34. Li YR, Keating BJ. Trans-ethnic genome-wide association studies: advantages and challenges of mapping in diverse populations. Genome Med. 2014;6:91.
Cite This Article
How to Cite
Download Citation
Export Citation File:
Type of Import
Tips on Downloading Citation
Citation Manager File Format
Type of Import
Direct Import: When the Direct Import option is selected (the default state), a dialogue box will give you the option to Save or Open the downloaded citation data. Choosing Open will either launch your citation manager or give you a choice of applications with which to use the metadata. The Save option saves the file locally for later use.
Indirect Import: When the Indirect Import option is selected, the metadata is displayed and may be copied and pasted as needed.
About This Article
Special Topic
Copyright
Data & Comments
Data

















Comments
Comments must be written in English. Spam, offensive content, impersonation, and private information will not be permitted. If any comment is reported and identified as inappropriate content by OAE staff, the comment will be removed without notice. If you have any queries or need any help, please contact us at [email protected].