fig2
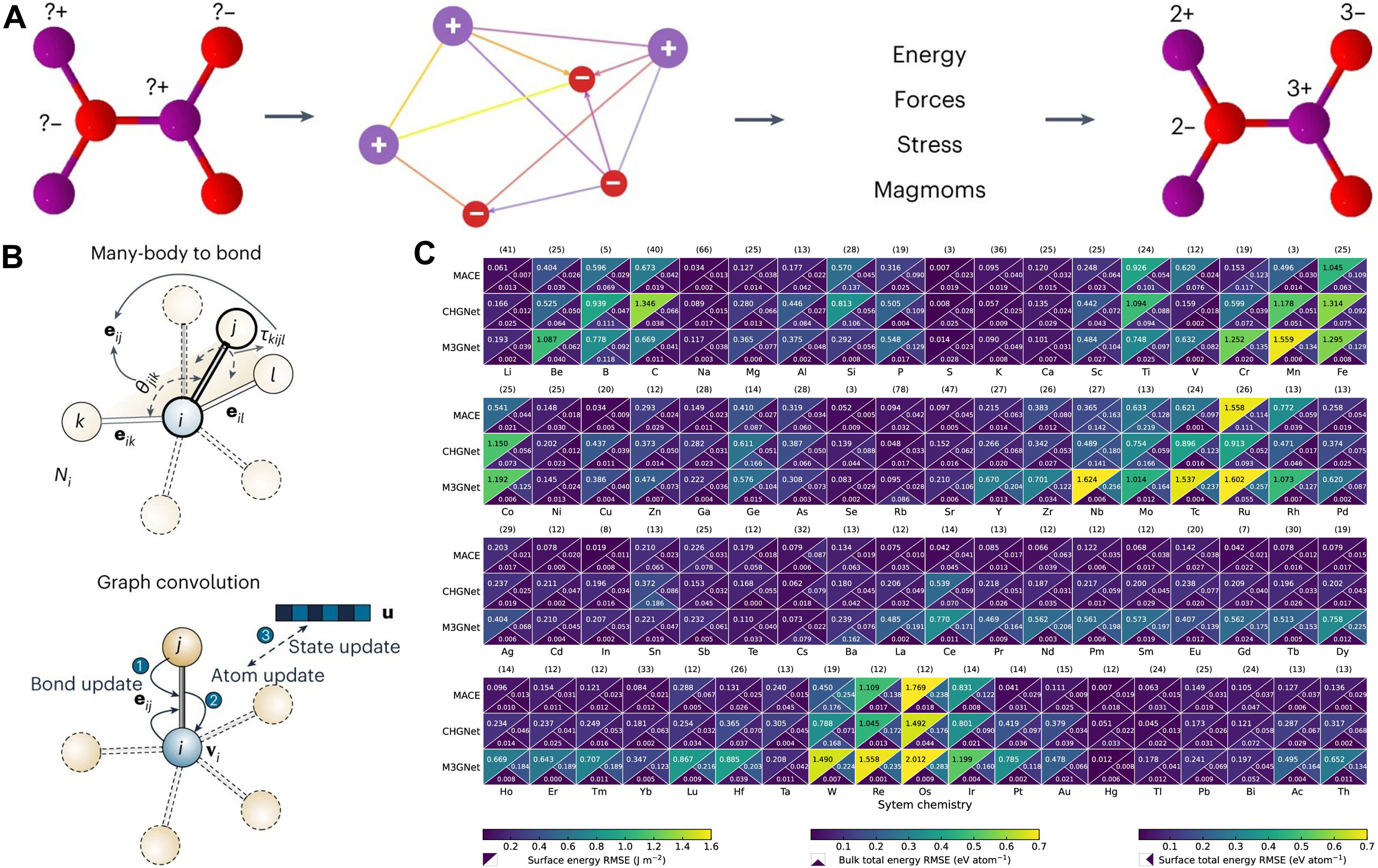
Figure 2. (A) CHGNet (dual graph, charge-informed): a crystal is represented by an atom graph (bond connectivity) and a bond graph (three-body/angular terms), yielding predictions of energy, forces, stress, magnetic moments, and a charge-decorated structure. Adapted with permission from Deng et al.[15] (CC BY 4.0); changes made: cropped to include only panel a; (B) M3GNet (many-body-to-bond): a graph convolution architecture that builds bond features with explicit angular terms (e.g., spherical harmonic features) and uses atomic coordinates and the lattice to compute stress. Adapted with permission from Chen & Ong[16] (© Springer Nature, 2022); change made: cropped to include only the rightmost “Many-body to bond” and “Graph convolution” panels; (C) Cross model benchmark (MACE, CHGNet, M3GNet): per element heatmaps report upper triangle: surface energy RMSE (J m-2); lower left: bulk total energy RMSE (eV atom-1); lower right: surface total energy RMSE (eV atom-1); numbers in parentheses give sample counts per element; lower values are better. Adapted with permission from Focassio et al.[18] (© American Chemical Society 2024); no changes made. CHGNet: Crystal Hamiltonian Graph neural Network; CC BY 4.0: Creative Commons Attribution 4.0 International; M3GNet: Materials 3-body Graph Network; MACE: Message-passing Atomic Cluster Expansion; RMSE: root-mean-square error.