Histone H3K18 lactylation and cisplatin resistance in bladder cancer: insights from single-cell transcriptomic analysis
Abstract
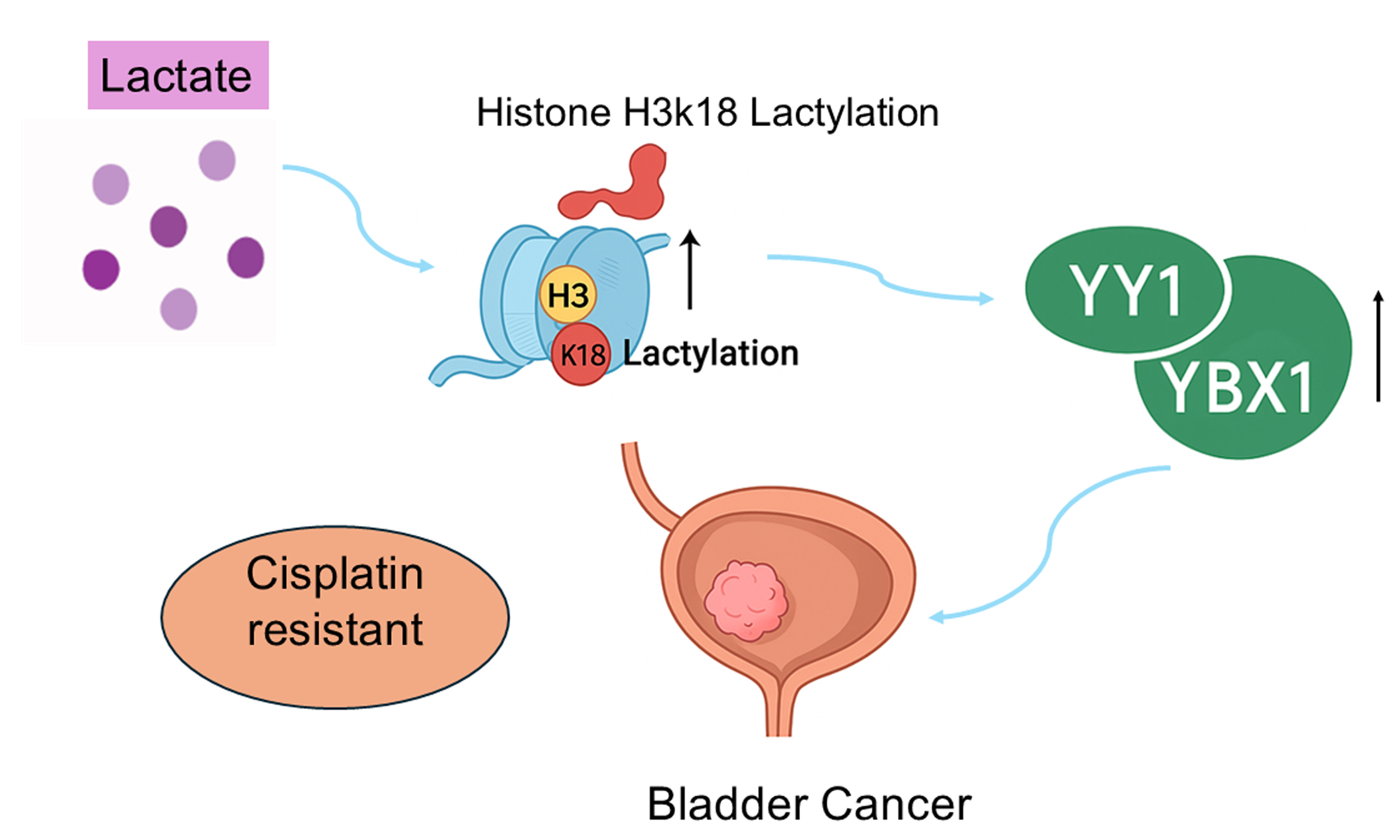
The study by Li et al., “Single-cell transcriptome analysis reveals the association between histone lactylation and cisplatin resistance in bladder cancer”, investigated how histone lactylation contributes to cisplatin resistance in bladder cancer (BCa). Using high-resolution single-cell lineage tracing, the authors identified a distinct subpopulation of BCa cells with enhanced glycolytic metabolism that exhibited significant cisplatin resistance. Further analyses showed significant enrichment of histone H3 lysine 18 lactylation (H3K18la) at the promoter regions of the transcription factors YY1 and YBX1, which promoted their expression and facilitated the development of a drug-resistant phenotype. This study was the first to combine single-cell omics with lactylation modification mechanisms, providing new insights into strategies for overcoming cisplatin resistance in BCa. Nonetheless, potential limitations of the study should be carefully considered.
Keywords
THE APPLICATION OF SINGLE-CELL TRANSCRIPTOME TECHNOLOGY IN BCA RESEARCH
In their article[1] published in Drug Resistance Updates, Li et al.[1] used single-cell transcriptomic analysis to comprehensively characterize the relationship between post-translational modifications and cisplatin resistance in bladder cancer (BCa). In recent years, single-cell RNA sequencing (scRNA-seq) has emerged as a powerful tool for resolving tumor heterogeneity[2]. In this study, the authors first identified tumor epithelial cell subpopulations linked to cisplatin resistance through bioinformatics analysis of BCa. They then further analyzed these subpopulations using the SCENIC and NetAct algorithms and identified a series of key transcription factors activated in the drug-resistant clusters, including YY1 and YBX1. By intersecting the genes regulated by YY1 and YBX1 with the marker genes of cisplatin-resistant epithelial subpopulations, they constructed a regulatory network encompassing DNA repair, apoptosis, and epithelial-mesenchymal transition (EMT). This analysis revealed that these transcription factors may drive cisplatin resistance in BCa by regulating multiple downstream drug resistance-related genes. Additionally, the authors inferred the differentiation trajectories of tumor cell subpopulations and analyzed the dynamic changes in key genes during tumor differentiation, providing a new perspective on the mechanisms of drug resistance.
HISTONE H3K18 LACTYLATION AND CISPLATIN RESISTANCE
Lactate, the final product of glycolysis, can form lactyl-CoA in the nucleus, thereby driving protein lactylation[3]. Li et al.[1] found that H3K18 lactylation (H3K18la) was significantly upregulated in resistant cells and demonstrated through ChIP-seq that H3K18la was enriched in the promoter regions of multiple genes. Notably, H3K18la was particularly enriched in the promoter regions of the transcription factors YY1 and YBX1, enhancing their transcriptional activation and expression, and ultimately driving the
TRANSCRIPTION FACTORS YY1 AND YBX1 AS KEY REGULATORS OF CISPLATIN RESISTANCE
Li et al.[1]identified YY1 and YBX1 as key transcriptional regulators of drug resistance. Both are multifunctional transcription factors implicated in drug resistance across various cancers[4,5]. YY1 is a versatile protein that not only regulates gene transcription but also participates in protein modification. Its overexpression has been closely llinked to chemotherapy tolerance in multiple tumor types[6]. Consistent with this, Li et al.[1] observed upregulated YY1 expression in resistant cells, which corresponded with H3K18la enrichment, suggesting that H3K18la may promote drug resistance by enhancing YY1 transcription.
YBX1, a member of the cold shock protein family, is recognized as a multifunctional driver of oncogenesis. Numerous studies have shown that YBX1 activates the transcription of genes related to proliferation, survival, and drug tolerance[7,8]. In the study by Li et al.[1], inhibition of glycolysis or disruption of histone lactylation not only diminished H3K18la levels but also impaired YBX1 activity, ultimately resensitizing BCa cells to cisplatin. These findings highlight YBX1 as a downstream effector that links metabolic reprogramming with epigenetic regulation, reinforcing its role in mediating drug resistance in BCa.
PROSPECTS FOR TRANSLATION AND RESEARCH LIMITATIONS
The study by Li et al.[1] offers a novel strategy for overcoming cisplatin resistance in BCa through
CONCLUSION
The single-cell transcriptomic study by Li et al.[1] innovatively revealed the key role of histone lactylation in cisplatin resistance in BCa and elucidated how lactate, a metabolic product, promotes drug resistance by regulating the expression of key transcription factors YY1 and YBX1. This discovery provides a new perspective on the mechanisms of tumor drug resistance and offers a theoretical basis for designing combined therapies that target metabolic-epigenetic pathways. Despite current limitations in experimental and clinical validation, this study provides important insights into BCa drug resistance and is expected to foster the development of more effective treatment regimens.
DECLARATIONS
Authors’ contributions
Literature review (equal), writing - original draft: Yang Z
Literature review (equal), writing - review and editing: Chen S
Availability of data and materials
Not applicable.
Financial support and sponsorship
None.
Conflicts of interest
Chen S is an Editorial Board member of Journal of Translational Genetics and Genomics. Chen S was not involved in any aspect of the editorial process for this manuscript, including reviewer selection, manuscript handling, or decision making. Yang Z declared that there are no conflicts of interest.
Ethical approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Copyright
© The Author(s) 2025.
REFERENCES
1. Li F, Zhang H, Huang Y, et al. Single-cell transcriptome analysis reveals the association between histone lactylation and cisplatin resistance in bladder cancer. Drug Resist Updat. 2024;73:101059.
2. Tirosh I, Suva ML. Cancer cell states: Lessons from ten years of single-cell RNA-sequencing of human tumors. Cancer Cell. 2024;42:1497-506.
3. Sahu V, Lu C. Metabolism-driven chromatin dynamics: molecular principles and technological advances. Mol Cell. 2025;85:262-75.
4. Li B, Xing F, Wang J, et al. YBX1 as a therapeutic target to suppress the LRP1-β-catenin-RRM1 axis and overcome gemcitabine resistance in pancreatic cancer. Cancer Lett. 2024;602:217197.
5. Yokoyama NN, Pate KT, Sprowl S, Waterman ML. A role for YY1 in repression of dominant negative LEF-1 expression in colon cancer. Nucleic Acids Res. 2010;38:6375-88.
6. Zhao L, Li R, Gan YH. Knockdown of Yin Yang 1 enhances anticancer effects of cisplatin through protein phosphatase 2A-mediated T308 dephosphorylation of AKT. Cell Death Dis. 2018;9:747.
7. Zhang X, An K, Ge X, et al. NSUN2/YBX1 promotes the progression of breast cancer by enhancing HGH1 mRNA stability through m5C methylation. Breast Cancer Res. 2024;26:94.
8. Wang X, Guo T, Niu L, et al. Engineered targeting OIP5 sensitizes bladder cancer to chemotherapy resistance via TRIP12-PPP1CB-YBX1 axis. Oncogene. 2024;43:2850-67.
9. Ge T, Gu X, Jia R, et al. Crosstalk between metabolic reprogramming and epigenetics in cancer: updates on mechanisms and therapeutic opportunities. Cancer Commun. 2022;42:1049-82.
Cite This Article
How to Cite
Download Citation
Export Citation File:
Type of Import
Tips on Downloading Citation
Citation Manager File Format
Type of Import
Direct Import: When the Direct Import option is selected (the default state), a dialogue box will give you the option to Save or Open the downloaded citation data. Choosing Open will either launch your citation manager or give you a choice of applications with which to use the metadata. The Save option saves the file locally for later use.
Indirect Import: When the Indirect Import option is selected, the metadata is displayed and may be copied and pasted as needed.
About This Article
Copyright
Data & Comments
Data

















Comments
Comments must be written in English. Spam, offensive content, impersonation, and private information will not be permitted. If any comment is reported and identified as inappropriate content by OAE staff, the comment will be removed without notice. If you have any queries or need any help, please contact us at [email protected].