The relationship between serum alkaline phosphatase levels and sepsis: observational and Mendelian randomization studies
Abstract
Aim: There is no definitive evidence to establish a causal relationship between elevated serum alkaline phosphatase (ALP) levels and sepsis susceptibility. This study employed observational and Mendelian randomization (MR) studies to investigate their potential correlation.
Methods: This observational study used data from the eICU Collaborative Research Database (eICU-CRD) and included adult patients who were admitted to the ICU for a single episode, with an ICU stay of ≥ 24 h, and serum ALP records obtained within 24 hours of admission. Based on three quantiles of ALP, participants were divided into three groups (Units/L): ALP ≤ 59, 60 ≤ ALP ≤ 86, and ALP ≥ 87. Multivariate logistic regression (MLR) explored the association between serum ALP levels and sepsis susceptibility in the general population and subgroups categorized by sex, age, blood lactate, and APACHE IV scores. Meanwhile, this study utilized genome-wide association studies (GWAS) data of serum ALP levels (Neale lab, n = 13,586,006) and sepsis (UK Biobank, n = 12,243,539) to conduct MR analysis to validate their causal relationship by the inverse variance weighted (IVW) method.
Results: The observational study comprised 4,458 patients. The sepsis susceptibility of the three groups was 11.1%, 12.5%, and 17.4%, respectively. MLR analysis revealed that sepsis susceptibility of the ALP ≥ 87 group was 1.748 times higher than the ALP ≤ 59 group (OR = 1.748, 95%CI: 1.399-2.183). There was a consistent correlation between them in sex, age, blood lactate, and the APACHE IV scores subgroup. MR study demonstrated a causal association between them (OR = 1.005, 95%CI: 1.002-1.008).
Conclusions: Elevated serum ALP levels are significantly associated with sepsis susceptibility, and there is a causal relationship between them.
Keywords
INTRODUCTION
Sepsis is characterized by a life-threatening organ dysfunction resulting from the host’s impaired response to infection[1]. According to the 2017 Global Burden of Disease (GBD) study, approximately 49 million individuals worldwide suffer from sepsis annually, leading to nearly 11 million deaths, which account for 19.7% of global mortality[2]. Despite significant advancements in medical technology, both the incidence and mortality rates of sepsis continue to rise, and survivors often experience a reduced quality of life[3]. Sepsis treatment costs are among the highest[4], and currently, no specific treatment is available. Therefore, it is crucial to identify risk factors that influence its onset and prognosis.
Alkaline phosphatase (ALP) is a membrane-bound glycoprotein that catalyzes the hydrolysis of phosphate esters under alkaline conditions. Most ALP in serum originates from the liver and bones, with minor contributions primarily from the kidneys and other tissues[5]. The typical range for ALP activity in adult serum is between 40 and 150 Units/L[6]. Recently, serum ALP levels have been used as predictors of various health conditions, including hepatobiliary disorders, sarcopenia, cardiovascular disease, peripheral arterial disease, metabolic syndrome, and multiple myeloma[7-12]. Additionally, alkaline phosphatase acts as an inflammatory mediator, similar to C-reactive protein (CRP). CRP has been identified as a novel risk marker for sepsis in patients undergoing outpatient maintenance hemodialysis[13]. Studies have consistently demonstrated a significant direct association between ALP and CRP, suggesting the potential for shared biological pathways[14,15]. However, due to confounding factors in observational studies, the causal relationship between serum ALP levels and inflammatory diseases, especially sepsis, remains to be further explored.
Mendelian randomization (MR) analysis is a promising epidemiological approach that utilizes genetic tools to assess causal relationships between exposure and outcomes. Compared with observational studies, MR studies are more effective in controlling for confounding factors and reverse causation, as genetic variants are independent of environmental risk factors and precede disease onset[16]. Genetic tools help identify biomarkers involved in clinical outcomes, disentangling correlation from causation[17,18]. MR analysis has identified causal relationships between various serum biomarkers and sepsis incidence, including serum lipid levels[19], serum iron levels[20], blood metabolite levels[21], and circulating cytokine levels[22,23].
With the advancement of genome-wide association studies (GWAS), it has become possible to investigate the causal relationship between serum ALP levels and sepsis incidence using a MR approach. In this study, we integrated observational analysis with two-sample MR analysis to explore the association and causal relationship between serum ALP levels and sepsis for the first time. Our aim is to provide robust evidence for early monitoring and intervention in sepsis.
METHODS
Retrospective cohort study
Data source and study design
This cohort study used the eICU Collaborative Research Database (eICU-CRD, version 2.0), a publicly accessible database maintained by the Laboratory for Computational Physiology at the Massachusetts Institute of Technology (MIT; Cambridge, MA, USA)[24]. eICU-CRD is a multicenter database containing de-identified data from 335 ICUs across 208 hospitals in the continental United States between 2014 and 2015. The dataset includes demographic characteristics, laboratory, radiology, and physiological data, medical histories, diagnoses based on the International Classification of Diseases, Ninth Edition (ICD-9) or Tenth Edition (ICD-10), treatment details, discharge status, and other relevant information. The safe harbor provision of the Health Insurance Portability and Accountability Act authorizes the release of the database. As all protected health information is de-identified, individual patient consent was not required. All authors were granted permission to access eICU-CRD.
Patients, independent variable, and outcome
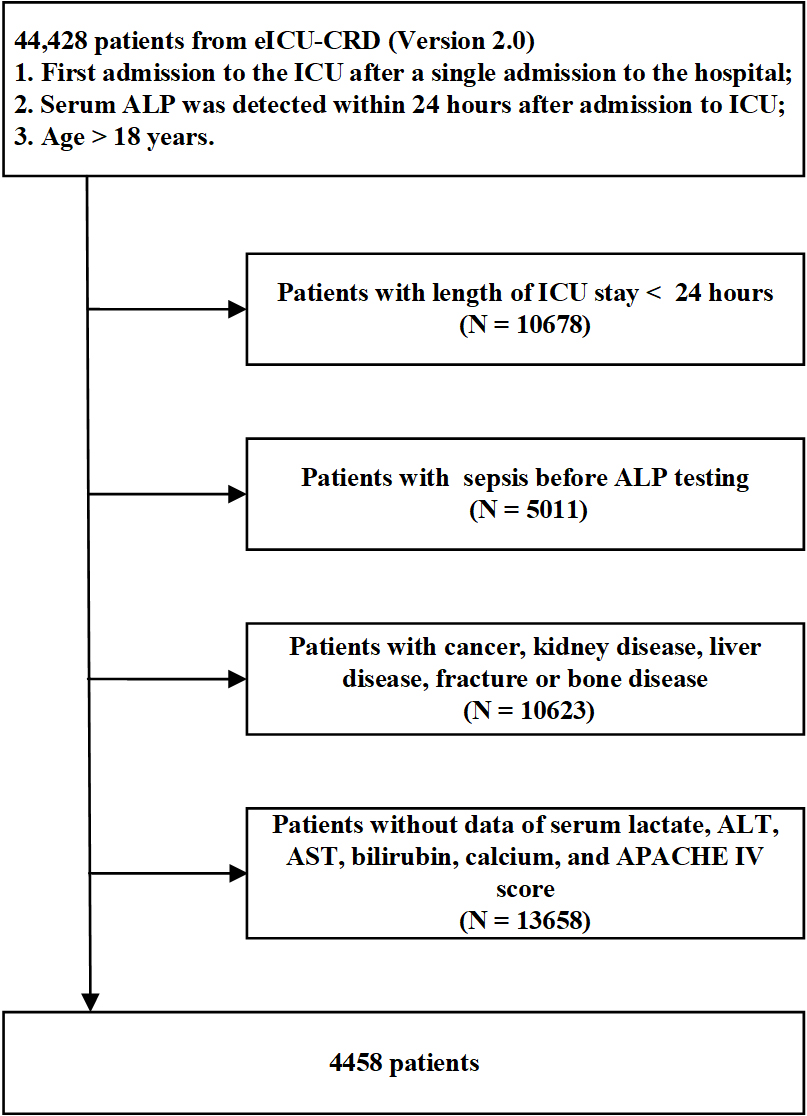
Patients were recruited from the eICU-CRD database. For individuals with multiple intensive care unit (ICU) admissions, only data from their first ICU admission were utilized, and the first recorded value was taken for patients with multiple laboratory tests. The inclusion criteria were as follows: (1) Patients with their first ICU admission during a single hospitalization; (2) Patients with documented serum ALP levels within 24 h of admission to the ICU; (3) Patients with age ≥ 18 years. The exclusion criteria were as follows: (1) Patients with a duration of ICU hospitalization less than 24 h; (2) Patients with confirmed sepsis prior to ALP testing; (3) Patients with certain conditions, including tumors, kidney diseases (such as nephritis, chronic kidney disease, and renal failure), liver diseases (such as hepatitis, cirrhosis, and liver failure), and musculoskeletal system diseases; (4) Patients without documented serum lactate, alanine aminotransferase (ALT), aspartate aminotransferase (AST), bilirubin, calcium levels, and the first Acute Physiology and Chronic Health Evaluation (APACHE) IV scores [Figure 1].
Figure 1. Flow diagram for patient recruitment. eICU-CRD: eICU collaborative research database; ICU: intensive care unit; ALP: alkaline phosphatase; ALT: alanine aminotransferase; AST: aspartate aminotransferase; APACHE: acute physiology and chronic health evaluation.
We extracted the following data for each patient from eICU-CRD: age, sex, ethnicity, the first APACHE IV scores, diseases diagnosed based on ICD-9 or ICD-10 during the first 24 h of ICU stay (diabetes, hypertension, coronary heart disease, respiratory failure, and disseminated intravascular coagulation), serum lactate, ALT, AST, bilirubin, calcium in the first 24 h after ICU admission, antibiotics treatment, invasive ventilation treatment, and sepsis diagnosed with ICD-9 or ICD-10 during ICU stay. Data were extracted using SAS version 9.4 (SAS Institute, Cary, NC, USA). The eICU-CRD (version 2.0) database records only APACHE IV scores and does not include APACHE II or SOFA scores. Therefore, APACHE IV scores are used to assess patient severity.
In this study, serum ALP levels were measured within 24 h of ICU admission. Specifically, we selected data with “chart time” - “intime” < 24 h based on the ALP detection time (“chart time”) and ICU entry time (“intime”) for further analysis. The outcome variable is the incidence of sepsis, defined explicitly as sepsis confirmed after admission to the ICU and ALP testing. The criteria for determining sepsis are based on the serum ALP detection time (“chart time”) and sepsis diagnosis time (“diagnosis time”) within 24 h of ICU admission. We selected data where “diagnosis time” - “chart time” > 0 h for the final study population.
Statistical analysis
The data of all patients were managed and statistically analyzed by SPSS 22.0 software. In descriptive analysis, Chi-square tests were utilized to compare groups for all count variables presented as frequency (percentage). The normality of all continuous variables was assessed using the Kolmogorov-Smirnov test and the histogram of normality test, revealing that none of the continuous variables adhered to a normal distribution. As a result, all continuous variables are presented as medians and inter-quartile ranges (IQRs), and group comparisons are conducted using the non-parametric Kruskal-Wallis H test.
The statistical software SPSS 22.0 was utilized for conducting univariate and multivariate logistic regression (MLR) analyses to examine the impact of serum ALP levels, categorized into three groups based on the third percentile, on the risk of sepsis in critically ill patients. Four models were built to show the modeling process in MLR analyses, which could support the stability of the association between serum ALP levels and the incidence of sepsis. Multivariable Model1 was adjusted for age, gender, ethnicity, and APACHE IV scores; Multivariable Model2 was adjusted for Multivariable Model1 + diabetes, hypertension, coronary heart disease, respiratory failure, and disseminated intravascular coagulation (DIC); Multivariable Model3 was adjusted for Multivariable Model2 + serum lactate, ALT, AST, bilirubin, and calcium; Multivariable Model4 was adjusted for Multivariable Model3 + antibiotics treatment and invasive ventilation treatment. Meanwhile, subgroup analysis was conducted to determine whether there were differences in the correlation between serum ALP levels and the risk of sepsis based on different APACHE IV scores, serum lactate levels, age, and gender. A two-sided P-value < 0.05 was considered statistically significant. For subgroup and sensitivity analyses involving multiple comparisons, we applied the false discovery rate (FDR) adjustment to minimize type I error.
Mendelian randomization study
Study design
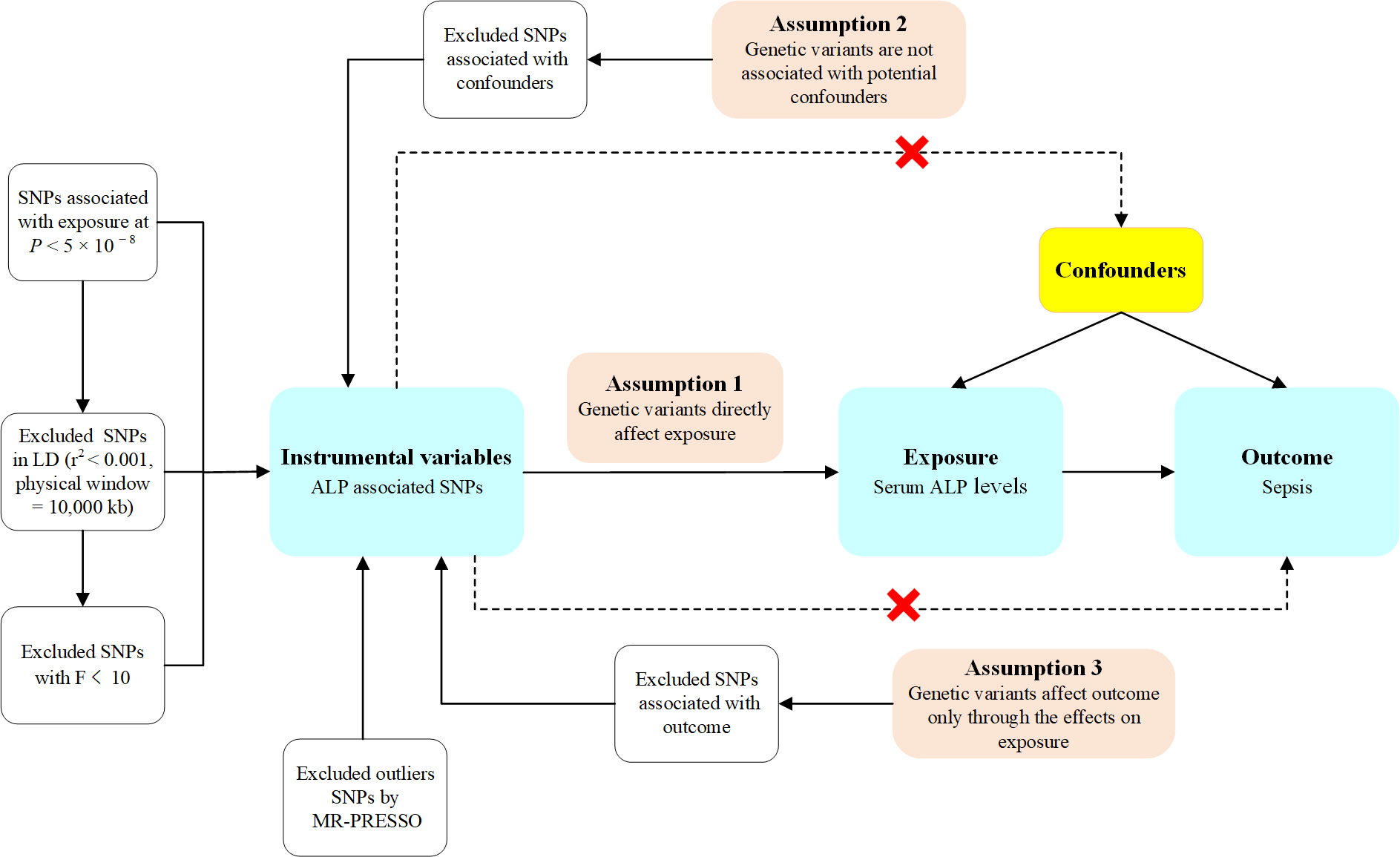
The study design for MR analysis is illustrated in Figure 2. To make genetic variants qualified as a valid instrument for causal inference, the MR approach we used must satisfy the following three core assumptions: (1) Genetic variants directly influence exposures; (2) genetic variants are not associated with potential confounders; and (3) genetic variants affect outcomes only via the effects on exposures[16]. Ethical approval and informed consent were obtained from participants for all original studies. As the data used in this study were publicly available, ethical approval and informed consent were not required according to the study design. Additionally, this study’s results were reported per the Strengthening the Reporting of Observational Studies in Epidemiology - Mendelian Randomization (STROBE-MR) guidance from 2021[25].
Figure 2. Overview of the design of the Mendelian randomization (MR) study. Illustration of the assumptions underpinning a MR analysis of the association between an exposure and an outcome. The arrows represent causal pathways. The dashed arrows represent potential causal associations between variables that would violate the assumptions of MR analysis. SNPs: Single nucleotide polymorphisms; LD: linkage disequilibrium.
GWAS data summary for exposures and outcomes
After searching the IEU OpenGWAS project (https://gwas.mrcieu.ac.uk/)[26], we selected two GWASs of European descent (Neale lab database and UK Biobank database) with the single-nucleotide polymorphisms (SNPs) of serum ALP levels and sepsis, respectively. Details of these GWASs are displayed in Supplementary Table 1. We used SNPs associated with serum ALP levels from the Neale lab study (OpenGWAS ID: ukb-d-30610_raw), including 13,586,006 SNPs. The study by UK Biobank was the largest GWAS for sepsis (OpenGWAS ID: ieu-b-4980) of European ancestry, including 11,643 cases of sepsis and 474,841 control subjects. Sepsis was defined based on the ICD-9 code and the ICD-10 code.
Selection of genetic instruments
Based on these criteria, SNPs were used as instrumental variables (IVs): (1) SNPs significantly correlated with serum ALP levels (P < 5 × 10-8); (2) To eliminate linkage disequilibrium (LD), SNPs were clumped based on LD threshold (r2 < 0.001) and distance (10,000 kb); (3) Except for SNPs present in the outcome GWAS data and proxy SNPs identified with proxies = TRUE, we removed SNPs not present in the outcome GWAS data; (4) SNPs without significant horizontal pleiotropy (MR-PRESSO global tests P > 0.05); (5) To avoid distortion of strand orientation or allele coding, we deleted palindromic SNPs (e.g., with A/T or G/C alleles); (6) SNPs associated with confounders were removed via the PhenoScanner website (https://ldlink.nci.nih.gov/?tab=ldtrait)[27]; (7) SNPs with F > 10. The F statistics for each SNP were computed using the following formula: F = (R2/k)/{[1-R2]/[n-k-1]}, where R2 is the proportion of risk factor variability explained by genotype, k is the number of instruments used in the model, and n is the sample size[28].
Statistical analysis
This study used inverse variance weighting (IVW) as the primary analysis method to infer a potential causal relationship between serum ALP levels and sepsis. The weighted median method and MR-Egger regression complement IVW for MR analysis. The odds ratio (OR) and 95% confidence interval (95%CI) were utilized to present the causal estimates, and P < 0.05 was deemed statistically significant. Heterogeneity was assessed according to Cochran’s Q test: if P ≥ 0.05, indicating no heterogeneity, a fixed-effects IVW model was applied. If P < 0.05, indicating heterogeneity, a random-effects IVW model was used when heterogeneity was acceptable[29,30]. Horizontal pleiotropy was examined utilizing the MR-Egger intercept test and the
RESULTS
Retrospective cohort study
Characteristics of study subjects
The present study ultimately recruited 4,458 patients from the eICU-CRD database, including 1,849 (41.5%) patients aged over 65 years, 2,241 (50.3%) females, and 3,499 (78.5%) Caucasians. The median (IQR)
Characteristics of the study subject
| Characteristics | Total (n = 4,458) | ALP ≤ 59 (n = 1,475) | 60 ≤ ALP ≤ 86 (n = 1,476) | ALP ≥ 87 (n = 1,507) | P |
| Age > 65, n (%) | 1,849 (41.5) | 619 (42.0) | 614 (41.6) | 616 (40.9) | 0.827 |
| Female, n (%) | 2,241 (50.3) | 681 (46.2) | 731 (49.5) | 829 (55.0) | < 0.001 |
| Ethnicity | 0.859 | ||||
| White, n (%) | 3,499 (78.5) | 1,152 (78.1) | 1,165 (78.9) | 1,182 (78.4) | |
| Other, n (%) | 959 (21.5) | 323 (21.9) | 311 (21.1) | 325 (21.6) | |
| APACHE IV scores, median (IQR) | 63 (46-83) | 62 (46-82) | 61 (45-81) | 65 (48-86) | < 0.001 |
| Diseases, n (%) | |||||
| Diabetes | 369 (8.3) | 102 (6.9) | 109 (7.4) | 158 (10.5) | < 0.001 |
| Hypertension | 393 (8.8) | 132 (8.9) | 146 (9.9) | 115 (7.6) | 0.091 |
| Coronary heart disease | 392 (8.8) | 136 (9.2) | 130 (8.8) | 126 (8.4) | 0.709 |
| Respiratory failure | 1,440 (32.3) | 439 (29.8) | 516 (35.0) | 485 (32.2) | 0.010 |
| DIC | 11 (0.2) | 7 (0.5) | 2 (0.1) | 2 (0.1) | 0.098 |
| Lactate, median (IQR) | 1.7 (1.1-2.7) | 1.8 (1.2-2.9) | 1.6 (1.1-2.6) | 1.7 (1.1-2.6) | < 0.001 |
| ALT, median (IQR) | 29 (17-57) | 24 (15-42) | 28 (17-50) | 38 (21-81) | < 0.001 |
| AST, median (IQR) | 37 (22-84) | 33 (20-67) | 34 (20-71) | 50 (26-117) | < 0.001 |
| Bilirubin, median (IQR) | 0.6 (0.4-1.0) | 0.6 (0.4-1.0) | 0.6 (0.4-0.9) | 0.7 (0.4-1.2) | < 0.001 |
| Calcium, median (IQR) | 8.1 (7.5-8.6) | 7.9 (7.3-8.5) | 8.2 (7.6-8.7) | 8.2 (7.7-8.7) | < 0.001 |
| Treatment, n (%) | |||||
| Antibiotics | 1,517 (34.0) | 470 (31.9) | 485 (32.9) | 562 (37.3) | 0.004 |
| Invasive ventilation | 2,617 (58.7) | 895 (60.7) | 899 (60.9) | 823 (54.6) | < 0.001 |
| Outcome, n (%) | |||||
| Incidence of sepsis | 610 (13.7) | 164 (11.1%) | 184 (12.5%) | 262 (17.4%) | < 0.001 |
The association between serum ALP levels and incidence of sepsis in all patients.
We categorized serum ALP levels into three groups: ALP ≤ 59 Units/L, 60 Units/L ≤ ALP ≤ 86 Units/L, and ALP ≥ 87 Units/L. In univariate analysis, there was no statistically significant difference in the incidence of sepsis between the 60 Units/L ≤ ALP ≤ 86 Units/L group and the ALP ≤ 59 Units/L group (OR, 1.138; 95%CI, 0.910-1.424). The incidence of sepsis in patients with ALP ≥ 87 Units/L group was 1.682 times higher than that in the ALP ≤ 59 Units/L group (OR, 1.682; 95%CI, 1.364-2.075). After adjustment for age, sex, ethnicity, APACHE IV scores, diabetes, hypertension, coronary heart disease, respiratory failure, DIC, serum lactate, ALT, AST, bilirubin, calcium, antibiotics treatment, and invasive ventilation treatment, the ALP ≥ 87 Units/L group was still associated with a higher risk for sepsis than the ALP ≤ 59 Units/L group (OR, 1.748; 95%CI, 1.399-2.183) [Table 2].
The association between serum ALP levels and incidence of sepsis
| Model | ALP ≤ 59 OR (95%CI) | 60 ≤ ALP ≤ 86 OR (95%CI) | ALP ≥ 87 OR (95%CI) |
| Univariable model | 1 (ref) | 1.138 (0.910-1.424) | 1.682 (1.364-2.075) |
| Multivariable Model 1 | 1 (ref) | 1.146 (0.915-1.436) | 1.635 (1.323-2.020) |
| Multivariable Model 2 | 1 (ref) | 1.122 (0.894-1.408) | 1.641 (1.325-2.033) |
| Multivariable Model 3 | 1 (ref) | 1.218 (0.968-1.533) | 1.803 (1.446-2.248) |
| Multivariable Model 4 | 1 (ref) | 1.204 (0.955-1.518) | 1.748 (1.399-2.183) |
Subgroup analyses
To further address the potential influence of different covariates on the results, subgroup analyses were conducted in patients classified by APACHE IV scores, serum lactate, sex, and age. The results show that the association between serum ALP levels and the incidence of sepsis is consistent in patients varied by the above covariates after adjustment for age, sex, ethnicity, APACHE IV scores, diabetes, hypertension, coronary heart disease, respiratory failure, DIC, serum lactate, ALT, AST, bilirubin, calcium, antibiotics treatment, and invasive ventilation treatment [Table 3].
The association between serum ALP levels and incidence of sepsis stratified by APACHE IV score, serum lactate, gender, and age
| Model | ALP ≤ 59 OR (95%CI) | 60 ≤ ALP ≤ 86 OR (95%CI) | ALP ≥ 87 OR (95%CI) |
| APACHE IV score * | |||
| ≤ 62 | 1 (ref) | 1.188 (0.816-1.731) | 2.038 (1.417-2.932) |
| > 62 | 1 (ref) | 1.215 (0.903-1.635) | 1.578 (1.189-2.095) |
| Serum lactate (mmol/L) § | |||
| < 2.0 | 1 (ref) | 1.128 (0.828-1.537) | 1.478 (1.094-1.997) |
| ≥ 2.0 | 1 (ref) | 1.228 (0.863-1.746) | 2.076 (1.488-2.896) |
| Sex † | |||
| Female | 1 (ref) | 1.282 (0.930-1.767) | 1.613 (1.188-2.191) |
| Male | 1 (ref) | 1.110 (0.791-1.558) | 1.973 (1.418-2.745) |
| Age (years) ‡ | |||
| ≤ 65 | 1 (ref) | 1.330 (0.965-1.833) | 2.009 (1.475-2.736) |
| > 65 | 1 (ref) | 1.077 (0.768-1.510) | 1.465 (1.058-2.031) |
Mendelian randomization study
Instrumental selection for serum ALP levels
According to the Neale lab database, 252 genetic variants were obtained as the IVs (P < 5 × 10-8). After a series of filtering steps, 184 SNPs were ultimately retained for use as IVs. Specifically, we excluded: 14 SNPs that did not exist in the GWAS of the outcome event, 16 SNPs with horizontal pleiotropy, 7 SNPs for being palindromic with intermediate allele frequencies, 25 SNPs associated with confounders (such as serum lipid levels, circulating cytokines levels, and blood metabolites levels), which were deleted using the PhenoScanner tool (https://ldlink.nci.nih.gov/?tab=ldtrait), and 6 SNPs with F ≤ 10. The detailed SNP filtering process is shown in Supplementary Table 2, and detailed information about 184 SNPs is presented in Supplementary Table 3.
Mendelian randomization analysis between serum ALP levels and sepsis risk
The IVW method suggested a significant association between serum ALP levels and the risk of sepsis (OR, 1.005; 95%CI, 1.002-1.008). The weighted median method provided little evidence to support the association between serum ALP levels and sepsis (OR, 1.007; 95%CI, 1.002-1.013). The MR-Egger analysis (OR, 1.006; 95%CI, 0.998-1.014) did not demonstrate a statistically significant relationship between serum ALP levels and the risk of sepsis. While the direction of effect was consistent with the weighted median method, this provides additional evidence for the validity of the IVW findings [Table 4]. In conclusion, the three mentioned causal inference methods suggest a positive causal association between genetically predicted serum ALP levels and the susceptibility to sepsis, wherein an elevation in serum ALP levels corresponds to an increased risk of sepsis. Additionally, the scatter plot of MR analysis results also suggests a causal association between serum ALP levels and the incidence of sepsis, with a positive correlation observed between the two variables [Supplementary Figure 1]. The forest plot of MR analysis results summarized the SNP-specific and overall MR estimates for the causal effects on sepsis using the separately selected SNPs associated with serum ALP levels [Supplementary Figure 2].
The Mendelian randomization analyses of the causality between serum ALP levels and incidence of sepsis
| Exposure | Outcome | Method | Nsnp | Bate | SE | P | OR (95%CI) |
| ukb-d-30610_raw | ieu-b-4980 | IVW | 184 | 0.005 | 0.002 | < 0.001 | 1.005 (1.002-1.008) |
| MR Egger | 184 | 0.006 | 0.004 | 0.180 | 1.006 (0.998-1.014) | ||
| Weighted median | 184 | 0.007 | 0.003 | 0.012 | 1.007 (1.002-1.013) |
Sensitivity analyses
Our sensitivity analysis included the leave-one-out analysis, pleiotropy test, and heterogeneity test. The leave-one-out analysis [Supplementary Figure 3] demonstrated that the lines of all instrumental variables consistently align on the same side of the invalid line (0), indicating that the removal of any SNP does not significantly impact the results. Therefore, the MR causal estimation results exhibit robustness. The P > 0.05 detected by the Cochrane Q method suggests no significant heterogeneity in SNPs [Supplementary Table 4]. Furthermore, the symmetrical distribution of genetic variation across various factors indicated by the funnel plot provides additional support for the absence of substantial heterogeneity among the selected SNPs
DISCUSSION
Sepsis is a common and frequently occurring condition in intensive care units, representing a major
However, the relationship between ALP and sepsis in critically ill patients remains underexplored in the current literature. To address this gap, we conducted a retrospective cohort study using the eICU database, which showed that the incidence of sepsis was 1.682 times higher in patients with ALP ≥ 87 U/L compared to those with ALP ≤ 59 U/L (OR, 1.682; 95%CI, 1.364-2.075). After adjusting for potential confounders, such as age, sex, race, APACHE IV score, diabetes, hypertension, coronary heart disease, respiratory failure, DIC, serum lactate, ALT, AST, bilirubin, calcium levels, antibiotic use, and invasive ventilation, the association remained robust. Specifically, patients with ALP ≥ 87 U/L had a significantly higher risk of sepsis than those with ALP ≤ 59 U/L (adjusted OR, 1.748; 95%CI, 1.399-2.183). This relationship persisted across subgroups stratified by APACHE IV scores, serum lactate levels, sex, and age, indicating the consistency and stability of the findings.
To further investigate the potential causal relationship at the genetic level, we performed MR analysis using summary-level data from the Neale Lab and UK Biobank. The results consistently supported a causal association between genetically predicted serum ALP levels and susceptibility to sepsis (OR, 1.005; 95%CI, 1.002-1.008). However, given the relatively small effect size (OR, 1.005; 95%CI, 1.002-1.008), we do not recommend using serum ALP as an independent predictor of sepsis incidence in critically ill patients in clinical practice.
Elevated serum ALP levels may contribute to sepsis through various pathophysiological mechanisms. Early studies have suggested that elevated ALP levels may lead to abnormal bone density, metabolic disturbances, renal insufficiency, and impaired vascular health, all of which could increase susceptibility to infection and sepsis[33]. Another potential mechanism is that elevated serum ALP levels may increase susceptibility to sepsis by mediating long-term chronic inflammation. An animal experiment demonstrated that serum ALP levels can enhance susceptibility to sepsis by mediating long-term chronic inflammation[34]. However, our study was unable to determine whether elevated ALP contributes to sepsis susceptibility through tissue damage or systemic inflammation. Further basic and clinical research is required to elucidate the mechanisms through which ALP influences sepsis risk.
This study also encountered certain limitations. First, this study adopts a retrospective design, which may have a selection bias. Second, in the retrospective cohort study, the independent variable is the individual’s serum ALP level within 24 hours of admission to the ICU. However, baseline serum ALP levels are lacking. It may be more accurate to predict the incidence of sepsis by relying on baseline ALP levels to assess changes in serum ALP levels after admission to the ICU. Third, the majority of participants in the retrospective cohort study were of Caucasian descent, and the population in the MR study was predominantly of European ancestry. While this enhances the robustness of causal inference within this population, it may limit the generalizability of our findings to other racial or ethnic groups. Future studies utilizing multi-ethnic GWAS data are necessary to validate these results in more diverse populations. Fourth, due to the limited availability of publicly accessible data on various subtypes of serum ALP, it poses a challenge to conduct observational and MR studies on the association between serum ALP subtypes and the risk of sepsis. Further experimental work is required to delineate the roles of specific ALP subtypes in sepsis pathogenesis, and large-scale prospective trials with standardized protocols are essential to establish ALP’s utility in routine clinical practice.
CONCLUSION
An observational study found that elevated serum ALP levels were significantly associated with an increased risk of sepsis. Additionally, two-sample MR analysis confirmed a causal relationship between serum ALP levels and the incidence of sepsis. Our findings suggest that elevated serum ALP may serve as a potential biomarker for sepsis risk; however, prospective studies are required to validate its predictive value and clinical utility across diverse populations and settings.
DECLARATIONS
Acknowledgments
The authors thank the researchers at the MIT Laboratory for Computational Physiology and their collaborating research groups for maintaining access to the eICU-CRD databases. The authors also acknowledge the IEU OpenGWAS project (https://gwas.mrcieu.ac.uk/) for providing summary-level GWAS data on serum ALP levels and sepsis. Gratitude is further extended to the participants and researchers involved in the Neale lab and the UK Biobank databases.
Authors’ contributions
Took responsibility for the integrity of the data, interpretation, and analysis: Wang G, Liu X, Li R, Li J
Contributed substantially to the study design, data interpretation, and writing of the manuscript: Liu X, Li R, Li J, Ren J, Zhang C, Xu X
Performed the statistical analysis and data synthesis: Wang G, Liu X, Li R, Li J, Ren J, Deng G, Zhang C, Xu X, Yuan Q, Gao X
All authors approved the final version of the manuscript.
Availability of data and materials
Retrospective cohort study data are available from the corresponding author upon reasonable request. The datasets used and/or analyzed during the current study are available from the corresponding author upon reasonable request. The data on serum ALP levels and sepsis were obtained from the IEU OpenGWAS project (https://gwas.mrcieu.ac.uk/).
Financial support and sponsorship
This work was supported by the Young Talent Support Program of Shaanxi Province of China.
Conflicts of interest
All authors declared that there are no conflicts of interest.
Ethical approval and consent to participate
This study was exempt from ethical approval as it involved only the analysis of publicly available summary-level data.
Consent for publication
Not applicable.
Copyright
© The Author(s) 2025.
Supplementary Materials
REFERENCES
1. Singer M, Deutschman CS, Seymour CW, et al. The third international consensus definitions for sepsis and septic shock (Sepsis-3). JAMA. 2016;315:801-10.
2. Rudd KE, Johnson SC, Agesa KM, et al. Global, regional, and national sepsis incidence and mortality, 1990-2017: analysis for the Global Burden of Disease Study. Lancet. 2020;395:200-11.
3. Huang M, Cai S, Su J. The Pathogenesis of sepsis and potential therapeutic targets. Int J Mol Sci. 2019;20:5376.
4. Rocheteau P, Chatre L, Briand D, et al. Sepsis induces long-term metabolic and mitochondrial muscle stem cell dysfunction amenable by mesenchymal stem cell therapy. Nat Commun. 2015;6:10145.
5. Singh SB, Lin HC. Role of intestinal alkaline phosphatase in innate immunity. Biomolecules. 2021;11:1784.
6. Han Y, Chen J, Li Z, Chen H, Qiu H. Recent progress and prospects of alkaline phosphatase biosensor based on fluorescence strategy. Biosens Bioelectron. 2020;148:111811.
7. Iluz-Freundlich D, Grubert Van Iderstine M, Uhanova J, Zhang M, Knowles C, Minuk GY. Low serum alkaline phosphatase levels in patients with chronic liver diseases: Possible contributions to disease pathogenesis. Clin Res Hepatol Gastroenterol. 2021;45:101694.
8. Lee JH, Cho AR, Lee YJ. Relationship between serum alkaline phosphatase and low muscle mass index among Korean adults: a nationwide population-based study. Biomolecules. 2021;11:842.
9. Kunutsor SK, Apekey TA, Khan H. Liver enzymes and risk of cardiovascular disease in the general population: a meta-analysis of prospective cohort studies. Atherosclerosis. 2014;236:7-17.
10. Cheung BM, Ong KL, Wong LY. Elevated serum alkaline phosphatase and peripheral arterial disease in the United States National Health and Nutrition Examination Survey 1999-2004. Int J Cardiol. 2009;135:156-61.
11. Kim JH, Lee HS, Park HM, Lee YJ. Serum alkaline phosphatase level is positively associated with metabolic syndrome: a nationwide population-based study. Clin Chim Acta. 2020;500:189-94.
12. Mirjačić Martinović K, Vuletić A, Tišma Miletić N, et al. Increased circulating monocyte MDSCs positively correlate with serum Interleukin-10 in metastatic melanoma patients. Innate Immun. 2023;29:37-44.
13. Katasako A, Sasaki S, Raita Y, et al. Association between serum alkaline phosphatase and bacteraemia in haemodialysis outpatients: a multicentre retrospective cross-sectional study. BMJ Open. 2022;12:e058666.
14. Cheung BM, Ong KL, Cheung RV, et al. Association between plasma alkaline phosphatase and C-reactive protein in Hong Kong Chinese. Clin Chem Lab Med. 2008;46:523-7.
15. Kerner A, Avizohar O, Sella R, et al. Association between elevated liver enzymes and C-reactive protein: possible hepatic contribution to systemic inflammation in the metabolic syndrome. Arterioscler Thromb Vasc Biol. 2005;25:193-7.
17. Cohen JC, Boerwinkle E, Mosley TH Jr, Hobbs HH. Sequence variations in PCSK9, low LDL, and protection against coronary heart disease. N Engl J Med. 2006;354:1264-72.
18. Li X, Hastie AT, Hawkins GA, et al. eQTL of bronchial epithelial cells and bronchial alveolar lavage deciphers GWAS-identified asthma genes. Allergy. 2015;70:1309-18.
19. Zeng L, Tang H, Chen J, et al. Causal association of lipoprotein-associated phospholipids on the risk of sepsis: a Mendelian randomization study. Front Endocrinol. 2023;14:1275132.
20. Hu Y, Cheng X, Mao H, Chen X, Cui Y, Qiu Z. Causal effects of genetically predicted iron status on sepsis: a two-sample bidirectional Mendelian randomization study. Front Nutr. 2021;8:747547.
21. Zhang Z, Yin Y, Chen T, et al. Investigating the impact of human blood metabolites on the Sepsis development and progression: a study utilizing two-sample Mendelian randomization. Front Med. 2023;10:1310391.
22. Zhi F, Ma JW, Ji DD, Bao J, Li QQ. Causal associations between circulating cytokines and risk of sepsis and related outcomes: a two-sample Mendelian randomization study. Front Immunol. 2024;15:1336586.
23. Fang W, Chai C, Lu J. The causal effects of circulating cytokines on sepsis: a Mendelian randomization study. PeerJ. 2024;12:e16860.
24. Pollard TJ, Johnson AEW, Raffa JD, Celi LA, Mark RG, Badawi O. The eICU Collaborative Research Database, a freely available multi-center database for critical care research. Sci Data. 2018;5:180178.
25. Skrivankova VW, Richmond RC, Woolf BAR, et al. Strengthening the reporting of observational studies in epidemiology using mendelian randomization: the STROBE-MR statement. JAMA. 2021;326:1614-21.
26. Hemani G, Zheng J, Elsworth B, et al. The MR-Base platform supports systematic causal inference across the human phenome. eLife. 2018;7:e34408.
27. Kamat MA, Blackshaw JA, Young R, et al. PhenoScanner V2: an expanded tool for searching human genotype-phenotype associations. Bioinformatics. 2019;35:4851-3.
28. Burgess S, Thompson SG. Bias in causal estimates from Mendelian randomization studies with weak instruments. Stat Med. 2011;30:1312-23.
29. Burgess S, Thompson SG. Interpreting findings from Mendelian randomization using the MR-Egger method. Eur J Epidemiol. 2017;32:377-89.
30. Bowden J, Del Greco M F, Minelli C, et al. Improving the accuracy of two-sample summary-data Mendelian randomization: moving beyond the NOME assumption. Int J Epidemiol. 2019;48:728-42.
31. Bowden J, Davey Smith G, Burgess S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol. 2015;44:512-25.
32. Zhang CY, Zhang HH, Zhao SL, et al. Clinical value of alkaline phosphatase on the surface membrane of neutrophils for prediction of bacteremia in patients with systemic inflammatory response syndrome. Diagn Microbiol Infect Dis. 2021;100:114105.
33. Park HM, Lee JH, Lee YJ. Positive association of serum alkaline phosphatase level with severe knee osteoarthritis: a nationwide population-based study. Diagnostics. 2020;10:1016.
Cite This Article
How to Cite
Download Citation
Export Citation File:
Type of Import
Tips on Downloading Citation
Citation Manager File Format
Type of Import
Direct Import: When the Direct Import option is selected (the default state), a dialogue box will give you the option to Save or Open the downloaded citation data. Choosing Open will either launch your citation manager or give you a choice of applications with which to use the metadata. The Save option saves the file locally for later use.
Indirect Import: When the Indirect Import option is selected, the metadata is displayed and may be copied and pasted as needed.
About This Article
Copyright
Data & Comments
Data


















Comments
Comments must be written in English. Spam, offensive content, impersonation, and private information will not be permitted. If any comment is reported and identified as inappropriate content by OAE staff, the comment will be removed without notice. If you have any queries or need any help, please contact us at [email protected].