Fabrication and evaluation of multiple pDNA-hydroxyapatite particles for transfection to HEK293 cells
Abstract
Aim: This study aimed to assess multiple marker-loaded plasmid DNA (pDNA)-calcium phosphate (CaP) particles (pDNA-CaP) to allow multiple tumor drug delivery for therapy. Despite its widespread use as an in vitro pDNA-CaP transfection, little attention has been paid to multiple delivery systems.
Methods: We optimized Ca:P ion ratios by varying ion concentrations in simulated body fluid solutions to induce sufficient protein expression. We examined the effect of the multiple marker-loaded pDNA-hydroxyapatite (HA) particles, including three markers of pDNA on the transfection to human embryonic kidney (HEK293) cells.
Results: The presence of three proteins inside the cells demonstrated successful transfection of multiple pDNAs when using the multiple plasmid-loaded (3-pDNA-HA) particles having 2.4 µm in size.
Conclusion: The 3-pDNA-HA showed effective transfection for HEK293 cells in comparison to 3 different single-pDNA-HA.
Keywords
Introduction
Transfection leads to the introduction of DNA, RNA, or oligonucleotides into eukaryotic cells. This process involves the uptake of extracellular molecules through the cell membrane into the cytoplasm and also into the nucleus. When DNA is brought into the nucleus, it can be incorporated into a cell’s genetic material and can induce the production of specific proteins[1]. Many vectors have been investigated for the purposes of transfection.
Hydroxyapatite [Ca10(PO4)6(OH)2), HA] particles may be effective for non-viral intracellular gene delivery or transfection and may be used for gene silencing. Plasmid DNA (pDNA) and RNA binding to HA particles occur through electrostatic interactions between Ca2+ in HA carriers and phosphate groups in pDNA or RNA[2].
An elegant investigation was conducted to demonstrate the mineral efficiency for pDNA release from calcium phosphate (CaP) mineral coatings[3]. The coatings were conducted in simulated body fluid (SBF) solutions of varying ion content and concentrations with pDNA. The composition of HA minerals led to varying mineral stability and, in turn, varying release rates of bound pDNA in simulated physiological environments[3-7]. The pDNA coated by CaP chemical exhibited feature sizes of 500 nm; decreasing their size led to high gene delivery. This was because of the ability of cells to endocytose the smaller particles, facilitating the release of DNA from CaP matrix. Furthermore, the efficiency of the gene transfer from the mineralized surface suggested that the amount of pDNA affected the gene expression[3,7].
In addition, HA are not prone to enzymatic degradation in physiological conditions, unlike organic or polymeric agents[8,9]. Thus, non-immunogenic responses, non-toxic degradation products, and pH-dependent solubility make pDNA-loaded HA nanoparticles (pDNA-HA) suitable for gene therapy[10,11].
Despite its widespread use in in vitro pDNA-CaP transfection, little attention has been paid to multiple delivery systems[12,13]. The single-factor delivery is oversimplified biology and has not been shown to be clinically efficacious in pathological states. In this study, we examined the effect of the multiple marker-loaded pDNA-HA particles, including three markers of pDNA in transfection of human embryonic kidney (HEK293) cells. This strategy is important as a methodology for treatment of pathological states[14].
We systematically investigated pDNA condensation in SBF, cation and counter-anion concentration (Ca:P ion ratios), and also precipitated agglomeration on the transfection efficiency for HEK293 cells. By optimization of these parameters, potent reverse transfection using gel substrate was demonstrated.
Methods
Materials
All chemicals were purchased from Nacalai Tesque, Kyoto. For the transfection experiments with cyclic pDNA, the pDNA mammalian expression vector encoding for green fluorescent protein (GFP, 227aa) was sourced from Monster Green® Fluorescent Protein hMGFP (Promega), encoded by an improved synthetic version of GFP gene originally cloned from the great star coral Montastrea cavernosa. The pDNA mammalian expression vectors for both red fluorescent protein (RFP, 226aa) and cyan fluorescent protein (CFP, 235aa) were purchased from Cosmobio Co. The pDNAs were propagated in the DH5a strain of E. coli bacteria and isolated by using a plasmid kit (QIAGEN® Plasmid Purification, QIAGEN) following the protocol, and stored in Milli-Q ultrapure water at -20 °C.
Preparation of pDNA-HA particles
Milli-Q ultrapure [18 MΩcm, total organic carbon (TOC) < 20 ppb] water was added into plastic beaker under magnetic stirring. The water in the beaker was heated to 36.5 °C. The mineralization modified-solution (mSBF) without CaCl2 (NaCl, 136.8 mmol/L; NaHCO3, 4.2 mmol/L; KCl, 3.0 mmol/L; K2HPO4·3H2O, 1.0 mmol/L; MgCl2·6H2O, 1.5 mmol/L; Na2SO4, 0.5 mmol/L; Tris base, 50 mmol/L) was adjusted to pH 7.40 using 1.0 mol/L of HCl.
pDNA-HA particles were prepared as follows: 35 mL aqueous solution with varying amounts of calcium chloride (20, 30, 40, 50, 160, 320, 500 mmol/L) was mixed with 35 mL aqueous solution of 200 µg/mL pDNA. The final pDNA concentration was 50 µg/mL. Furthermore, 70 µL of varying ions concentration of mSBF was added and incubated for 30 min to be grown as HA sediment on pDNA molecules. The ion concentrations are listed in Table 1. This dispersion was used in the transfection experiment.
Final ions concentration of calcium chloride-pDNA solution mixed with different ions concentration of mSBF
| Modified SBF | Added CaCl2 (mmol/L) | Final ions concentration (mmol/L) | pDNA (μg/mL) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Na+ | K+ | Ca2+ | Mg2+ | Cl− | HCO3− | HPO42− | SO42− | |||
| SBF | - | 142.0 | 5.0 | 2.5 | 1.5 | 147.8 | 4.2 | 1.0 | 0.5 | - |
| 2SBF | 20 | 142.0 | 5.0 | 5.0 | 1.5 | 152.8 | 4.2 | 1.0 | 0.5 | 50 |
| 30 | 142.0 | 5.0 | 7.5 | 1.5 | 157.8 | 4.2 | 1.0 | 0.5 | 50 | |
| 40 | 142.0 | 5.0 | 10 | 1.5 | 162.8 | 4.2 | 1.0 | 0.5 | 50 | |
| 50 | 142.0 | 5.0 | 12.5 | 1.5 | 167.8 | 4.2 | 1.0 | 0.5 | 50 | |
| 160 | 142.0 | 5.0 | 40 | 1.5 | 222.8 | 4.2 | 1.0 | 0.5 | 50 | |
| 320 | 142.0 | 5.0 | 80 | 1.5 | 302.8 | 4.2 | 1.0 | 0.5 | 50 | |
| 500 | 142.0 | 5.0 | 125 | 1.5 | 392.8 | 4.2 | 1.0 | 0.5 | 50 | |
| 3SBF | 20 | 142.0 | 6.0 | 5.0 | 1.5 | 152.8 | 4.2 | 1.5 | 0.5 | 50 |
| 30 | 142.0 | 6.0 | 7.5 | 1.5 | 157.8 | 4.2 | 1.5 | 0.5 | 50 | |
| 40 | 142.0 | 6.0 | 10 | 1.5 | 162.8 | 4.2 | 1.5 | 0.5 | 50 | |
| 50 | 142.0 | 6.0 | 12.5 | 1.5 | 167.8 | 4.2 | 1.5 | 0.5 | 50 | |
| 160 | 142.0 | 6.0 | 40 | 1.5 | 222.8 | 4.2 | 1.5 | 0.5 | 50 | |
| 320 | 142.0 | 6.0 | 80 | 1.5 | 302.8 | 4.2 | 1.5 | 0.5 | 50 | |
| 500 | 142.0 | 6.0 | 125 | 1.5 | 392.8 | 4.2 | 1.5 | 0.5 | 50 | |
| 4SBF | 20 | 142.0 | 7.0 | 5.0 | 1.5 | 152.8 | 4.2 | 2.0 | 0.5 | 50 |
| 30 | 142.0 | 7.0 | 7.5 | 1.5 | 157.8 | 4.2 | 2.0 | 0.5 | 50 | |
| 40 | 142.0 | 7.0 | 10 | 1.5 | 162.8 | 4.2 | 2.0 | 0.5 | 50 | |
| 50 | 142.0 | 7.0 | 12.5 | 1.5 | 167.8 | 4.2 | 2.0 | 0.5 | 50 | |
| 160 | 142.0 | 7.0 | 40 | 1.5 | 222.8 | 4.2 | 2.0 | 0.5 | 50 | |
| 320 | 142.0 | 7.0 | 80 | 1.5 | 302.8 | 4.2 | 2.0 | 0.5 | 50 | |
| 500 | 142.0 | 7.0 | 125 | 1.5 | 392.8 | 4.2 | 2.0 | 0.5 | 50 | |
For multiple marker-loaded pDNA-HA particles, the pDNA-HA particles containing 66.7 µg/mL of each pDNA (GFP, RFP and CFP, total of 200 µg/mL) were prepared as described above and were designated as 3-pDNA-HA.
As a control for transfection, the standard pDNA-CaP particles were prepared as follows: 35 mL aqueous solution containing of 250 mmol/L calcium chloride was mixed with 35 mL aqueous solution of 80 or 200 µg/mL pDNA. The final pDNA concentration was 50 µg/mL. In addition, 70 mL of HEPES buffered saline solution (2 × HBS: NaCl, 280 mmol/L: Na2HPO4, 1.5 mmol/L: HEPES, 50 mmol/L, pH = 7.10 ± 0.01)[15] was added and incubated for 30 min for growing CaP sediment on pDNA molecules (designated as pDNA-CaP). The ion concentrations are listed in Table 2. This dispersion was used as a control.
Ions concentration of pDNA-CaP preparation
| Added CaCl2 (mmol/L) | Final ions concentration (mmol/L) | pDNA (μg/mL) | |||
|---|---|---|---|---|---|
| Na+ | Ca2+ | Cl− | HPO42− | ||
| 500 | 141.5 | 125 | 390 | 0.75 | 50 |
Lipofectamine-mediated transfection (as a positive control) was conducted with the same procedure after preparing pDNA-lipofectamine complexes at a weight of 1:5 according to the protocol provided by the manufacturer (L3000, Invitrogene).
Characterization
Morphology was observed via field emission scanning electron microscope (FE-SEM: SU6600, Hitachi Ltd.). The operated accelerating voltage was 15 kV and the specimens were coated with a thin layer of gold and palladium (Au/Pd 6:4) with a thickness of ~20 nm on cupper grids with 200 mesh size.
The surface charge characteristics of pDNA-HA particles in ultrapure water were determined by electrophoresis at 25 °C (Zetasizer Nano ZS, Malvern Instruments, UK) by the technique of laser Doppler anemometry. The method involved dialysis (MWCO 12000-14000, Fisher Scientific) against deionized water for 12 h to remove free ions in dispersion and adjusting the pH 7.4 using dilute NaOH. All measurements were performed in triplicate and were averaged to obtain the final value.
The average diameter of the particles was measured by dynamic light scattering (DLS) using a Zetasizer Nano ZS (wavelength = 532 nm). The dynamic information could be retrieved by examining the autocorrelation function g(t) of the time-dependent intensity[16].
Cell culture and transfection
HEK293 cells were purchased from the American Type Culture Collection (ATCC). The HEK293 cell line has been extensively used an expression tool for recombinant proteins since it was generated in the early 70s by transformation of cultures of normal human embryonic kidney cells with sheared adenovirus 5 DNA in Alex van der Eb’s laboratory in the Netherlands. The current commercially available source of HEK293 cells are from the original transformation by Graham et al.[17].
Before cell seeding, a 96-well plate was uniformly coated with a 20 µg/mL solution (50 µL) of fibronectin (Fn) (F1141, Sigma Aldrich). Solution was aspirated, and plates were air-dried to enhance cell binding affinity arising from alteration in the structure of the adsorbed Fn[18].
HEK293 cells were cultured in high-glucose Dulbecco’s modified Eagle’s medium (DMEM, Life Technologies) supplemented with 10% fetal bovine serum (FBS, Life Technologies) and 1% antibiotic-antimycotic mixture (NacalaiTesque) in an atmosphere of 5% CO2 and 95% relative humidity at 37 °C. For transfection experiments, HEK293 cells were seeded in 96-well plates at 1.0 × 104 cells/cm2 in 100 µL of media and were cultured for 24 h. Then, 20 µL of the pDNA-HA dispersion was added to well. After incubation for 24 h, the transfection efficiency with pDNA-HA particles was calculated by the green, red, and cyan fluorescent cells per well area viewed under a fluorescent microscope (BIOREVO BZ-9000, KEYENCE Co.) with 470-nm excitation and a 525-nm emission band pass filter for GFP, a 553-nm excitation with a 570-nm emission band pass filter for RFP, and a 400-nm excitation with a 495-nm emission band pass filter for CFP. The fluorescent cells were analyzed by ImageJ software.
After exposure of HEK293 cells to pDNA-HA particles having various Ca:P ion ratios for 24 h, the transfection medium was replaced by fresh cell culture medium with HBSS (Hank’s balanced salt solution without Phenol Red, Wako Pure Chemical Industries, Ltd., Japan). A 10 µg/mL solution of Hoechst33342 (Life Technologies) in PBS was added for labeling nuclei to visualize the cells. Transfection efficiency was calculated as the ratio of the cells in which GFP and/or RFP and/or CFP were expressed divided by the total number of cells.
Cell viability was determined by WST-8 assay (Dojindo) according to the standard protocol. After exposure of HEK293 cells to particles for 24 h, 200 µL of WST-8 solution was added to a working solution (the cell culture medium of the transfected cells) and was incubated for 30 min in an atmosphere of 5% CO2 and 95% relative humidity at 37 °C. The WST-8 colorimetric test measured the activity of intracellular dehydrogenase activity that is proportional to living cells. The optical density was read on a SpectraMax® Plus384 (Molecular Devices Co.) at 450 nm for absorbance and at 650 nm to subtract background absorbance.
Reverse transfection
Collagen solutions were prepared by mixing solutions at 0 °C of 7:2:1 Collmatrix I-A/minimum essential medium (MEM) (10X) without NaHCO3/sterile reconstitution buffer (260 mmol/L of NaHCO3, 50 mmol/L of NaOH, and 200 mmol/L of HEPES [2-[4-(2-hydroxyethyl)piperazin-1-yl]ethane-sulfonic acid] (Nitta Gelatin Inc., Japan), yielding a homogeneous solution. Then, the collagen solution was added onto 96-well plates and was heated at 37 °C for 30 min to obtain the pDNA-HA particles-incorporated collagen gels. The particles were embedded on the collagen gel by centrifugation at 1000 rpm to fix the particles. The wells were washed gently several times with HBSS to remove the unembedded particles, prior to reverse transfection with HEK293 cells. The cells were seeded on the surface of the collagen gels with thickness of ~200 µm at a density of 4.0 × 104 cells/cm2.
Statistics
Statistical analysis was performed using Student’s t-test. Significance was considered at a probability of P < 0.05.
Results
Characterization of pDNA-HA particles
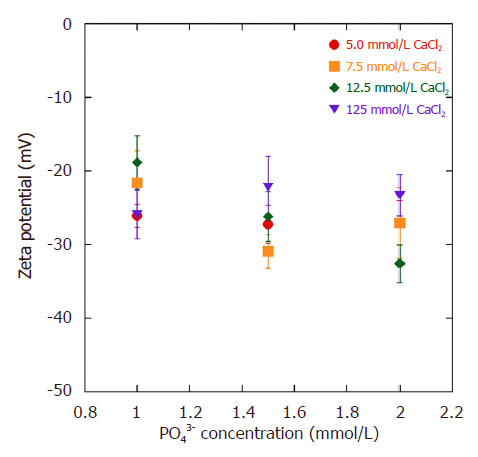
The pDNA-HA nanoparticles were generated by the electrostatic interaction between negatively charged phosphate (PO2-) groups along the backbone of the pDNA and Ca2+ that deposited calcium ions to interact with phosphate ions (PO43-) in the SBF to form HA. The presence of layers can be demonstrated by monitoring the zeta potential that changes from negative to positive when HA is on the outside and pDNA (-46 ± 0.7 mV) is on the inside, as demonstrated in our previous paper[19]. The zeta potentials of the pDNA-HA particles at pH 7.4 exhibited negative values (-17 to -33 mV) [Figure 1]. For 125 mmol/L CaCl2 loading condition, the zeta potentials increased only moderately from -27 to -23 mV, indicating dense HA layers including cations, (e.g., Ca2+, CaOH+) on the particle surfaces.
Figure 1. Zeta potential variation of pDNA-HA particleson different CaCl2 and PO43- ions concentrationat pH 7.4. Results are expressed as mean and SD (n = 3). pDNA: plasmid DNA; HA: hydroxyapatite
Cytotoxicity depends on the charge on the surface of the particles. Negatively-charged particles at pH 7.40 showed lower unfavorable effects on cell viability because of the negatively-charged cell membrane (about -20 mV) that plays an important role in separating the cytoplasm from the outside environment and modulating the movement of the particles in and out of the cell.
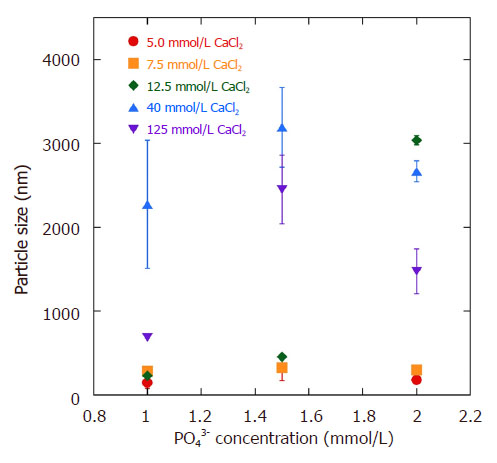
The particle size of pDNA-HA from DLS measurement is presented in Figure 2. The size of the low CaCl2 concentrations (5.0-12.5 mmol/L) were around 140 to 330 nm and the polydispersity index (PDI) for high PO43- concentration particles was between 0.13 and 0.16, indicating good monodispersity of the particles. It is possible to prevent aggregate formation at high values of zeta potential (about -30 mV)[20]. For high CaCl2 loading conditions (40-125 mmol/L), aggregate formation was observed in both 3SBF and 4SBF at high PO43- concentration. The particle size increased to ~3 µm, accompanied by high PDI, around 0.3 to 0.5.
Figure 2. Particle size of pDNA-HA vs. CaCl2 and PO43- ions concentration. Results are expressed as mean and SD (n = 3). pDNA: plasmid DNA; HA: hydroxyapatite
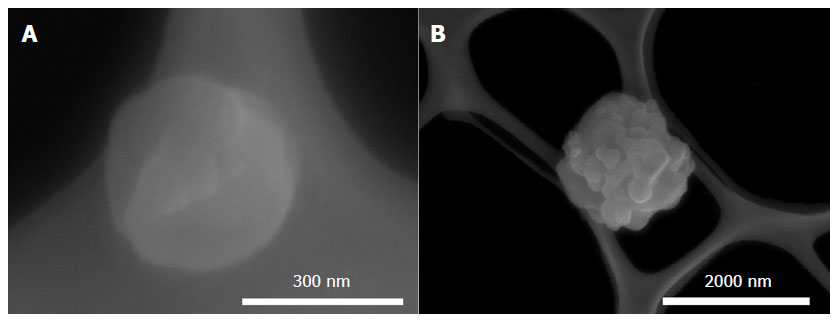
Figure 3 shows the morphological feature of pDNA-HA nanoparticles as revealed by FE-SEM imaging. The discrete particle exhibited very smooth surfaces with sizes of ~300 nm [Figure 3A]. The aggregate formation consisted of many discrete particles during the collection after biomineralization was also observed, in which the particle size increased to ~2 µm [Figure 3B].
Figure 3. FE-SEM images showing pDNA-HA nanoparticles prepared by 4SBF and CaCl2 of 125 mmol/L: (A) discrete particle and (B) aggregate formation. pDNA: plasmid DNA; HA: hydroxyapatite; SBF: simulated body fluid
In addition, the PDI of pDNA-HA particle (~0.25) fabricated by the ion concentration for CaCl2 of 12.5 mmol/L and PO43- of 1.5 mmol/L did not increase much, but the zeta potential decreased from greater than -20 mV. This finding indicated that the large surface charge disturbed the aggregation, causing a charge repulsive effect[20].
Transfection of pDNA-HA particles
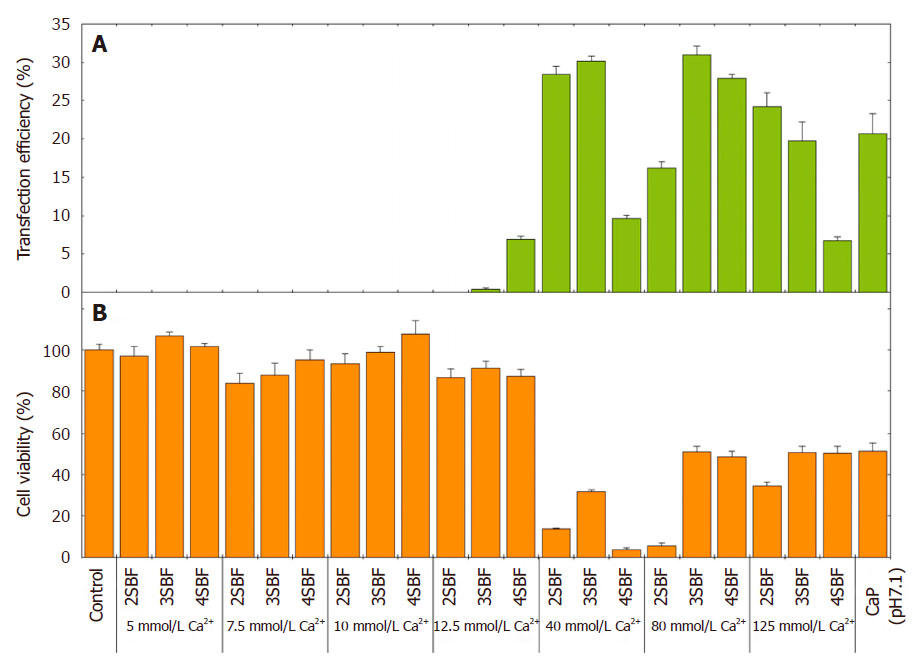
The transfection efficiency of GFP positive HEK293 cells at various mSBF concentrations with various CaCl2 loading conditions is shown in Figure 4. Beyond 12.5 mmol/L of CaCl2, the transfection efficiency improved to 30% with increasing CaCl2 concentrations in each mSBF. The Ca2+ ion concentration appeared to predominate for the gene transfection. By contrast, low CaCl2 loading (≤ 12.5 mmol/L) showed less transfection accompanied by high cell viability of near 100%, compared to untreated cells (100% viability). The gene transfection efficiency of pDNA-HA particles was around 30% at a particle size of 2.4 µm, comparable to the standard pDNA-CaP (~20%) that had large particle size (over 3.4 µm).
Figure 4. (A) Gene transfection efficiency and (B) cell viability of GFP positive HEK293 cells at different mSBF concentrations with different CaCl2 loading conditions. pDNA-CaP (at pH 7.1) as a standard control is shown in comparison. Results are expressed as mean and SD (n = 4). pDNA: plasmid DNA; CaP: calcium phosphate; mSBF: mineralization modified-solution; SBF: simulated body fluid
Transfection efficiency of multiple pDNA-HA particles
HEK293 cells transfected with pDNA-HA (CaCl2 loading of 125 mmol/L and 2SBF) were found to have least 6-fold more green fluorescence compared to cells transfected with lipofectamine-L3000 [Supplementary Figure 1]. This result was much in agreement with that of the pDNAs loaded on HA particles that were efficiently taken up by the cells, confirming the significantly better transfecting agent as compared to lipofectamine with the same amount of pDNA (50 µg/mL). As mentioned above, advances of pDNA-HA prompted us to conduct a multiple transfection (co-expression) study for pDNA-HA particles, while more study appears necessary to establish multiple delivery systems[12,13].
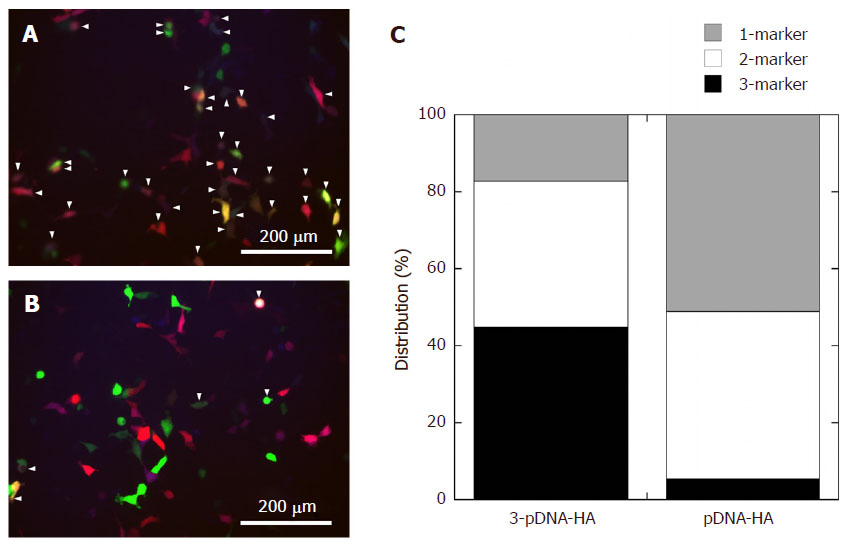
For 3-pDNA-HA (CaCl2 loading of 125 mmol/L and 2SBF), incubation of HEK293 cells expressing the marker proteins, GFP, RFP and CFP with HA particles and co-localization analysis are shown in Figure 5A. Four types of multiple transfections were analyzed by co-localization based on GFP/RFP/CFP-, GFP/RFP-, GFP/CFP-, and RFP/CFP-marker proteins, that were viewed under various emission band pass filters for each color. The cells expressing multiple fluorescence by overlaying green, red and cyan were observed (marked with the arrowheads in Figure 5A) following transfection of 3-pDNA-HA. The 3-pDNA-HA gave multiple transfection efficiency of more than 40% for three marker proteins of GFP/RFP/CFP and up to 38% for two markers [Figure 5C]. This feature indicated the existence of three proteins inside the cells, suggesting successful transfection of multiple pDNAs when using the 3-pDNA-HA. The cell viability was preserved (40% of cells survived) at concentration of CaCl2 loading of 125 mmol/L and 2SBF.
Figure 5. Representative fluorescent images of HEK293 cells incubated with (A) 3-pDNA-HA and (B) three different single marker-loaded pDNA-HAs. The arrowheads indicate the cells with expressing multiple fluorescence; (C) multiple transfection efficiency for each marker. pDNA: plasmid DNA; HA: hydroxyapatite
By contrast, for HEK293 cells transfected with three different single marker-loaded pDNA-HA particles, co-administration of these pDNA-HA particles, i.e., GFP-, RFP-, and CFP-markers, showed more diffuse distribution of the multiple transfection, based on three different fluorescences [Figure 5B]. In addition, the majority of the highly fluorescent GFP- and/or RFP-positive HEK293 cells were seen after transfection with pDNA-HA particles, indicating that the three markers were not fully transfected into cells. In this case, the transfection efficiencies of the co-administration of single marker-loaded pDNA-HA showed 50% for single marker-loaded pDNA-HA and ~44% for multiple transfection of two marker proteins. The multiple transfection of three marker proteins were significantly lower (~5%) [Figure 5C].
Reverse transfection
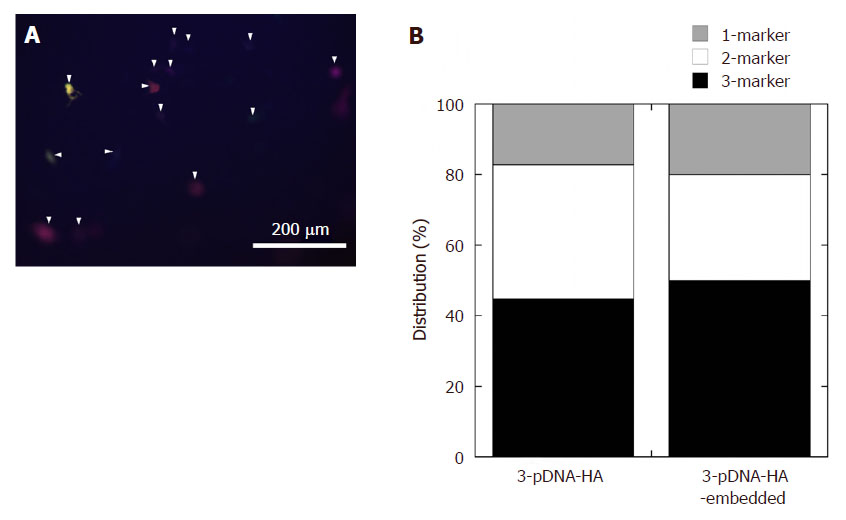
As shown in Figure 6A, significantly higher transfection levels were observed 1 day after incubation on the 3-pDNA-HA-embedded gel than that with administration of 3-pDNA-HA [Figure 5A] with same amount of pDNA (50 µg/mL).
Figure 6. (A) Representative fluorescent images of HEK293 cells incubated on 3-pDNA-HA-embedded gel. The arrowheads indicate the cells with expressing multiple fluorescence; (B) comparison of transfection efficiency for each marker between multiple transfections and reverse multiple transfection. pDNA: plasmid DNA; HA: hydroxyapatite
Discussion
The concern about particle size for transfection efficiency was addressed. No correlation between zeta potential and GFP expression was evident. For pDNA-HA with particle size 200-300 nm, gene transfection efficiencies were relatively lower compared to those having large particle size (about 2.5 µm). This behavior was not consistent with the optimum particle size for endocytosis, where the limited particle size is about 20-200 nm[21]. However, a report demonstrated that larger particles are much preferred due to the concentration of pDNA on the cell surface by sedimentation[22]. This correlated with phagocytosis as the particle entry mechanism, in which the large particle size was dominant.
Recently, researchers have commented that the addition of calcium chloride before transfection experiment promoted effective endocytosis and subsequent gene expression[23].
We tried to determine the release and transfection of pDNA-HA from a collagen gel substrate. The formation of larger aggregates cannot be easily internalized into cells via the endocytotic process, resulting in fewer transfected cells in this experimental system. However, there was a similar phenomenon at reverse multiple markers transfection. That is to say, the multiple transfection efficiency of three marker proteins was still higher regardless of the incubation system, as shown in Figure 6B.
In conclusion, we successfully prepared pDNA-HA particles fabricated with varying ion concentrations of SBF (Ca:P ion ratios) to investigate the transfection efficiency of the pDNA-HA particles to human embryonic kidney (HEK293) cells. Beyond 12.5 mmol/L of CaCl2, the transfection efficiency of pDNA-HA particles was around 30%, at a particle size of 2.4 µm, and was comparable to the standard pDNA-CaP (~20%) that have large particle size over 3.4 µm. Moreover, compared with that of lipofectamine, the pDNAs loaded on HA particles were efficiently taken up by the cells, confirming the presence of a significantly better transfecting agent.
We also examined the fabrication of 3-pDNA-HA nanoparticles, including three markers of pDNA and investigated the effect of multiple marker-loaded pDNA-HA particles on the transfection to the cells. We demonstrated three proteins inside the cells, suggesting successful transfection of multiple pDNAs using the 3-pDNA-HA.
As shown by reverse multi-transfection via 3-pDNA-HA-embedded gel substrate, the 3-pDNA-HA particles were very efficiently taken up through the cell membrane into the cytoplasm and into the nucleus. Therefore, the integration of pDNA-HA nanoparticles with loading of multiple pDNAs into tissue engineering may provide clinical relevance of drug delivery systems for therapy and for regeneration of functional tissues.
Declarations
Author’s contributions
Contributed to the initiating idea and performed most of the experiments: Sasaki R, Takeshita T, Okamoto M, Hirano M
Supervised the research: Okamoto M
Participated in writing the manuscript: all authors
Data source and availability
The data presented is original and obtained in our laboratory. It is available upon reasonable request to the corresponding author.
Financial support and sponsorship
This study was supported by the TTI Grant (Special Research Project: FY2014-2015).
Conflicts of interest
There are no conflicts of interest.
Patient consent
Not applicable.
Ethics approval
Experiments were performed on commercial HEK293 cell line. In accordance with the guideline of the Bioethics Committee of the Toyota Central R&D Labs, ethics approval is not required on cells purchased in the ATCC.
Copyright
© The Author(s) 2018.
REFERENCES
1. McNeil SE, Perrie Y. Gene delivery using cationic liposomesa. Expert Opin Ther Pat. 2006;16:1371-82.
2. Chen W, Lin M, Lin P, Tasi P, Chang Y, Yamamoto S. Studies of the interaction mechanism between single strand and double-strand DNA with hydroxyapatite by microcalorimetry and isotherm measurements. Colloids Surf A. 2007;295:274-83.
3. Shen H, Tan J, Saltzman WM. Surface-mediated gene transfer from nanocomposites of controlled texture. Nat Mater. 2004;3:569-74.
4. Shea LD, Smiley E, Bonadio J, Mooney DJ. DNA delivery from polymer matrices for tissue engineering. Nat Biotechnol. 1999;17:551-4.
5. Choi S, Murphy WL. Sustained plasmid DNA release from dissolving mineral coatings. Acta Biomater. 2010;6:3426-35.
6. Cheng B, Cai W, Yu J. DNA-mediated morphosynthesis of calcium carbonate particles. J Colloid Interface Sci. 2010;352:43-9.
7. Sun B, Yi M, Yacoob CC, Nguyen HT, Shen H. Effect of surface chemistry on gene transfer efficiency mediated by surface-induced DNA-doped nanocomposites. Acta Biomater. 2012;8:1109-16.
8. Cheung H, Lau K, Lu T, Hui D. A critical review on polymer-based bioengineered materials for scaffold development. Compos Part B Eng. 2007;38:291-300.
9. Habraken WJ, Zhang Z, Wolke JG, Grijpma DW, Mikos AG, Feijen J, Jansen JA. Introduction of enzymatically degradable poly(trimethylene carbonate) microspheres into an injectable calcium phosphate cement. Biomaterials. 2008;29:2464-76.
10. Kester M, Heakal Y, Sharma A, Robertson GP, Morgan TT, Altinoğlu Eİ, Tabaković A, Parette MR, Rouse S, Ruiz-Velasco V, Adair JH. Calcium phosphate nanocomposite particles for in vitro imaging and encapsulated chemotherapeutic drug delivery to cancer cells. Nano Lett. 2008;8:4116-21.
11. Morgan TT, Muddana HS, Altinoglu EI, Rouse SM, Tabaković A, Tabouillot T, Russin TJ, Shanmugavelandy SS, Butler PJ, Eklund PC, Yun JK, Kester M, Adair JH. Encapsulation of organic molecules in calcium phosphate nanocomposite particles for intracellular imaging and drug delivery. Nano Lett. 2008;8:4108-15.
12. Yuen WW, Du NR, Chan CH, Silva EA, Mooney DJ. Mimicking nature by codelivery of stimulant and inhibitor to create temporally stable and spatially restricted angiogenic zones. Proc Natl Acad Sci U S A. 2010;107:17933-8.
13. De la Riva B, Sánchez E, Hernández A, Reyes R, Tamimi F, López-Cabarcos E, Delgado A, Evora C. Local controlled release of VEGF and PDGF from a combined brushite-chitosan system enhances bone regeneration. J Control Release. 2010;143:45-52.
14. Browne S, Pandit A. Multi-modal delivery of therapeutics using biomaterial scaffolds. J Mater Chem B. 2014;2:6692-707.
15. Zhang Q, Mochalin VN, Neitzel I, Hazeli K, Niu J, Kontsos A, Zhou JG, Lelkes PI, Gogotsi Y. Mechanical properties and biomineralization of multifunctional nanodiamond-PLLA composites for bone tissue engineering. Biomaterials. 2012;33:5067-75.
16. Nishida Y, Domura R, Sakai R, Okamoto M, Arakawa S, Ishiki R, Salick MR, Turng L. Fabrication of PLLA/HA composite scaffolds modified by DNA. Polymer. 2015;56:73-81.
17. Graham FL, Smiley J, Russell WC, Nairn R. Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J Gen Virol. 1977;36:59-74.
18. Stephansson SN, Byers BA, García AJ. Enhanced expression of the osteoblastic phenotype on substrates that modulate fibronectin conformation and integrin receptor binding. Biomaterials. 2002;23:2527-34.
19. Takeshita T, Matsuura Y, Arakawa S, Okamoto M. Biomineralization of hydroxyapatite on DNA molecules in SBF: morphological features and computer simulation. Langmuir. 2012;29:11975-81.
20. Doostmohammadi A, Monshi A, Salehi R, Fathi HF, Golniya Z, Daniels UA. Bioactive glass nanoparticles with negative zeta potential. Ceram Int. 2011;37:2311-6.
21. Xiang SD, Scholzen A, Minigo G, David C, Apostolopoulos V, Mottram PL, Plebanski M. Pathogen recognition and development of particulate vaccines: does size matter? Methods. 2006;40:1-9.
Cite This Article
How to Cite
Download Citation
Export Citation File:
Type of Import
Tips on Downloading Citation
Citation Manager File Format
Type of Import
Direct Import: When the Direct Import option is selected (the default state), a dialogue box will give you the option to Save or Open the downloaded citation data. Choosing Open will either launch your citation manager or give you a choice of applications with which to use the metadata. The Save option saves the file locally for later use.
Indirect Import: When the Indirect Import option is selected, the metadata is displayed and may be copied and pasted as needed.
About This Article
Copyright
Data & Comments
Data













Comments
Comments must be written in English. Spam, offensive content, impersonation, and private information will not be permitted. If any comment is reported and identified as inappropriate content by OAE staff, the comment will be removed without notice. If you have any queries or need any help, please contact us at [email protected].