An exploratory in silico analysis of bacteriocin gene clusters in the urobiome
Abstract
Background: The role of the urobiome in health and disease remains an understudied area compared to the rest of the human microbiome. Enhanced culturing techniques and next-generation sequencing technologies have identified the urobiome as an untapped source of potentially novel antimicrobials. The aim of this study was to screen the urobiome for genes encoding bacteriocin production.
Methods: The genomes of 181 bacterial urobiome isolates were screened in silico for the presence of bacteriocin gene clusters using the bacteriocin mining tool BAGEL4 and secondary metabolite screening tool antiSMASH7.
Results: From these isolates, an initial 263 areas of interest were identified, manually annotated, and evaluated for potential bacteriocin gene clusters. This resulted in 32 isolates containing 80 potential bacteriocin gene clusters, of which 72% were identified as class II, 13.75% as class III, 8.75% as class I, and 5% as unclassified bacteriocins.
Conclusion: Overall, 53 novel variants were discovered, including nisin, gassericin, ubericin, and colicins.
Keywords
INTRODUCTION
The human microbiome and its role in health and disease have been at the forefront of scientific research in recent times[1-4]. To date, bacterial communities from numerous body sites have been screened, both in silico and in vitro, for antimicrobial compounds[5-11]. Recent advances in metaculturomics and metagenomic sequencing have led to the discovery and characterisation of the urobiome[12,13], which represents a relatively understudied environment in terms of the diversity and novelty of bacteriocins encoded by this microbial community.
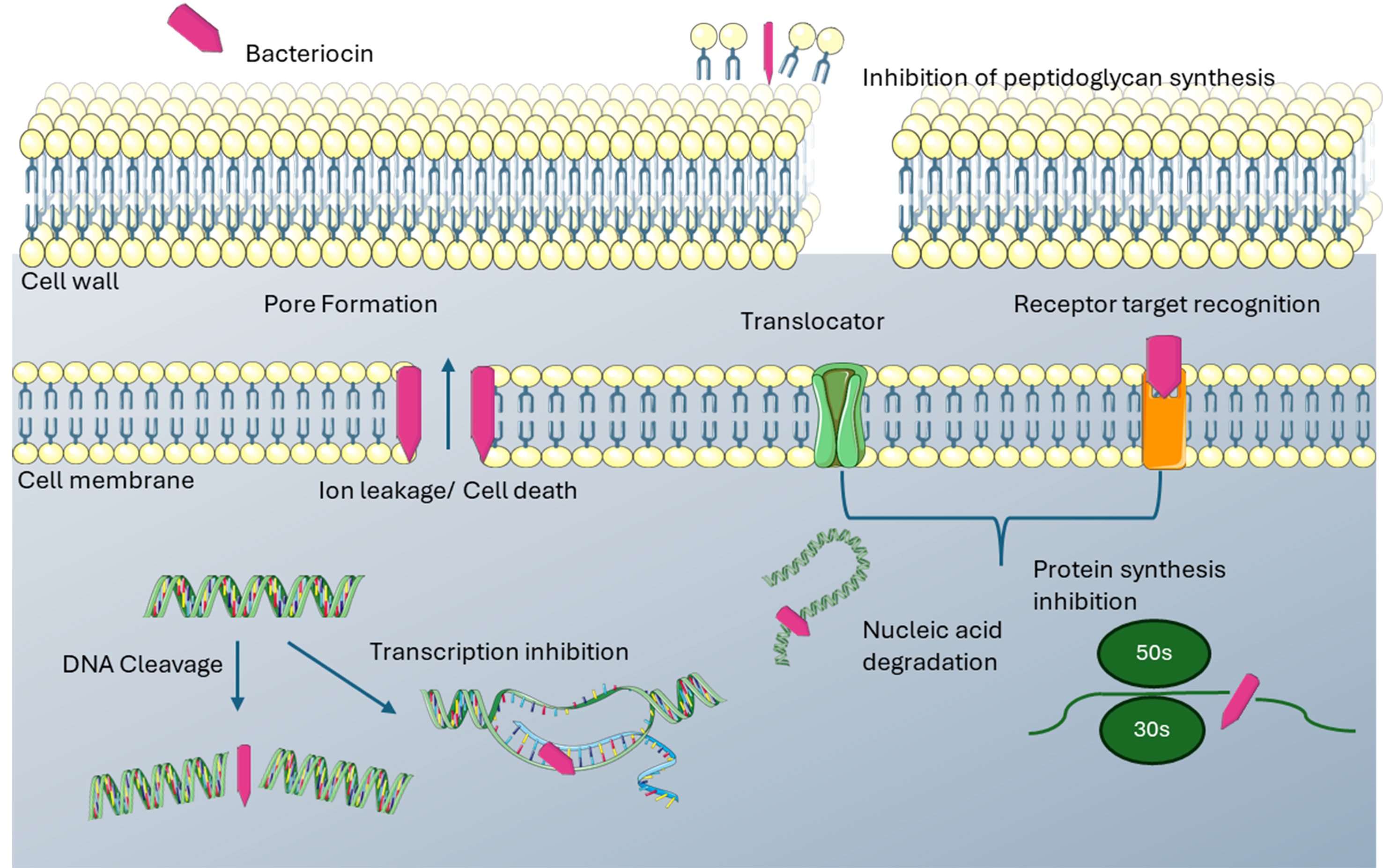
Bacteriocins are classified as ribosomally synthesised antimicrobial peptides, which are produced by bacteria as a defence mechanism against other bacteria present in the same environment[5,14,15]. Bacteriocins can display both narrow- and broad-spectrum bactericidal activity but are usually most effective against bacteria that are closely related to the producer strain[6,8,16-18]. While some bacteriocins are produced by Gram-negative bacteria[19], the majority of bacteriocins characterised to date are produced by Gram-positive, lactic acid bacteria[20-22]. Bacteriocins have been divided into three classes: class I, also known as lantibiotics, are characterised based on the presence of the amino acid lanthionine or methyllanthionine as a result of post-translational modifications. The primary mode of action of class I bacteriocins is targeting the cell membrane[14,20]. Class II bacteriocins are smaller, thermostable peptides that can be further categorised into five subclasses[23]. They are classed as broad range antimicrobials and act by forming pores in the cell membrane. Class III bacteriocins are larger, heat-sensitive peptides that cause bacterial cell lysis[5,14,20]. In previous years, food preservation and other applications in the food industry were the primary focus of bacteriocin research[15]. More recently, this focus has shifted to antimicrobial resistance, and strategies to improve the treatment and control of antibiotic-resistant infections[15], mainly centred on in vivo animal studies[24]. Bacteriocins have numerous desirable traits as antimicrobials, which make them particularly attractive alternatives to antibiotics, including low toxicity, high potency, and, most importantly, the ability to be effectively bioengineered[18,23]. Furthermore, a narrower activity spectrum than conventional antibiotics significantly reduces undesirable collateral damage to the commensal microbiota[2,11,14].
In silico screening of bacterial genomes for bacteriocin production has significantly reduced both the time and cost of culture-based approaches for bacteriocin discovery[25], with gene mining tools such as BAGEL4 and antiSMASH7[26,27] enabling the rapid identification of bacteriocin gene clusters. BAGEL4 scans the bacterial genome for putative bacteriocin open reading frames (ORFs). It searches for the structural bacteriocin gene, but also takes advantage of the common structure of bacteriocin operons and scans the surrounding ORFs for possible accessory genes that encode immunity, transport, regulation, and modification proteins[25,28,29]. AntiSMASH7, on the other hand, uses set “rules” that identify core biosynthetic functions present within a genomic region to create a biosynthetic gene cluster (BGC). AntiSMASH7 combines different profile hidden Markov model “rules” to identify 81 different BGC types[27].
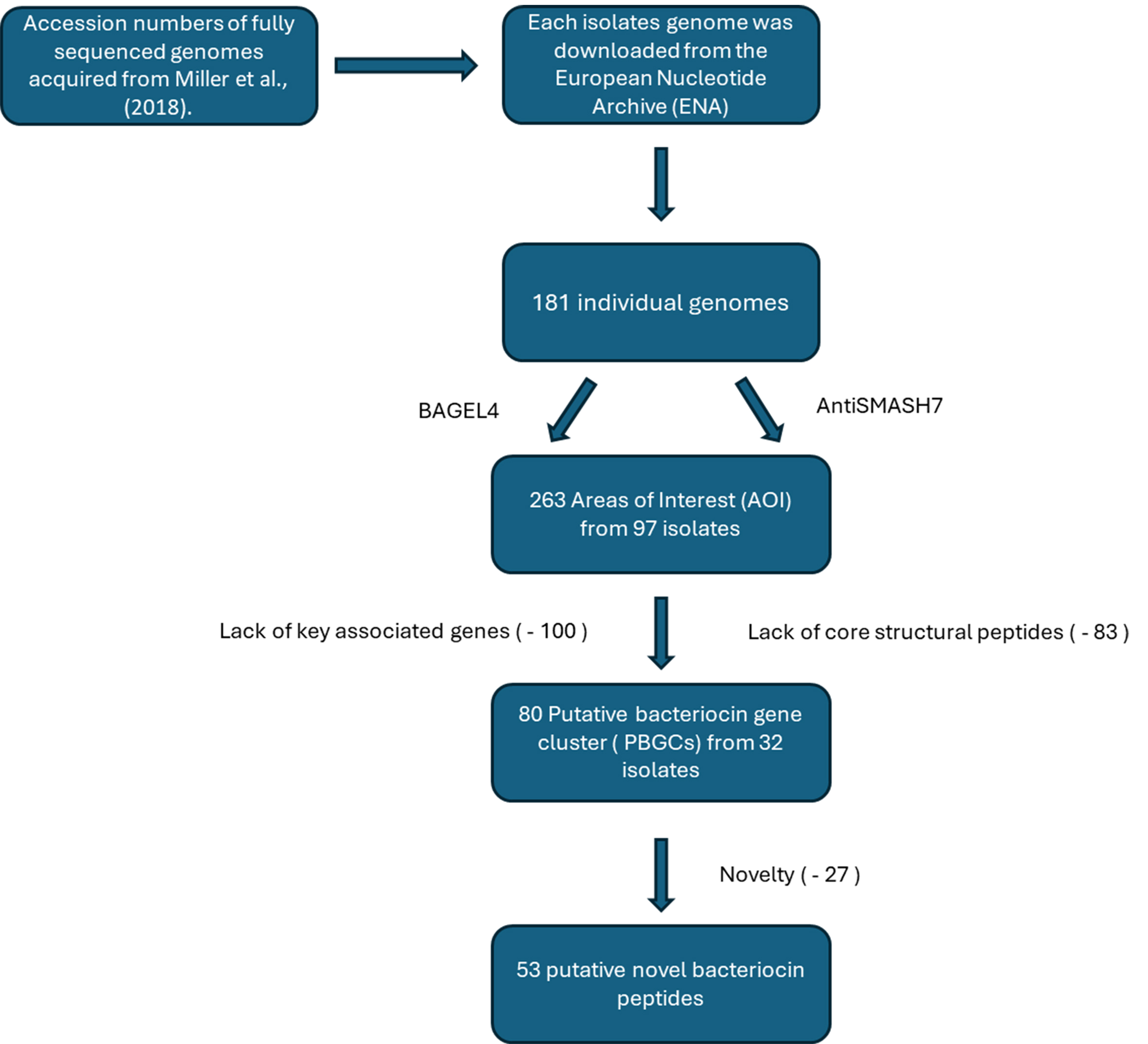
The current study is, to the best of our knowledge, the first to screen the urobiome for genes encoding bacteriocin production. Herein, we screened 181 bacterial isolates previously isolated from the bladder[30], with the primary objective to identify novel bacteriocin clusters in the urobiome [Figure 1]. Initially, 263 putative bacteriocin gene clusters were identified, highlighting the potential of the urobiome to host a diversity of bacteriocin producers.
METHODS
Data collection
The 181 fully sequenced genomes examined in this study were from urinary bacterial isolates, collected via catheter, previously isolated, sequenced, and assembled by Miller-Ensminger et al. (2018)[30]. Accession numbers were obtained [Supplementary Data], and each individual genome was downloaded from the European Nucleotide Archive (ENA) (https://www.ebi.ac.uk/ena/browser/home) in FASTA format.
Initial screening
The bacteriocin mining tool BAGEL4 (http://bagel4.molgenrug.nl/index.php)[26] was used in combination with antiSMASH7 (https://antismash.secondarymetabolites.org/#!/start)[27] to identify and analyse putative bacteriocin gene clusters (PBGCs), using default parameters.
Further analysis of individual gene clusters
Areas of interests (AOIs) identified by BAGEL4, predicted to be bacteriocins or associated with bacteriocin production, were investigated using BLASTP (https://blast.ncbi.nlm.nih.gov/Blast.cgi?PROGRAM=blastp&PAGE_TYPE=BlastSearch&LINK_LOC=blasthome)[31]. An AOI was considered to be a PBGC if it contained a structural core peptide and if it was surrounded by the key associated genes previously described in the literature, such as immunity, transport, leader cleavage, and a modification gene for post-translationally modified peptides[9,29]. To determine the degree of novelty of the PBGCs, the amino acid sequences for bacteriocin production were aligned against their closest characterised homologues, as indicated by BLASTP using the sequence alignment tool EMBL-EBI EMBOSS Needle (https://www.ebi.ac.uk/Tools/psa/) using the Needleman-Wunsch algorithm. Novelty was described when a difference of two or more amino acids was identified in the predicted bacteriocin sequence compared to previously characterised bacteriocins[10]. All amino acid sequences of the surrounding accessory proteins displayed > 95% identity to their predicted proteins, unless otherwise stated.
RESULTS
In silico screening for putative bacteriocin gene clusters
This study screened 181 bacterial urobiome isolates[30] cultured from catheterised urine samples. Catheterised urine samples reduced the risk of cross-contamination from surrounding microbiomes (urethra, skin, vagina, etc.), and, as such, represent true urobiome/bladder isolates[32]. The initial screening using BAGEL4 and antiSMASH7 resulted in the identification of 263 AOIs [Supplementary Table 1]. BAGEL4 identifies the presence of AOIs within the genomes; however, this does not necessarily translate into functional peptide production for reasons including mutations, regulation, or target specificity.
In total, 263 AOIs were identified from 97 isolates across 35 genera, with 54 of the isolates predicted to produce more than one putative bacteriocin [Supplementary Table 1]. Further analysis revealed that 83 of the AOIs lacked a core structural peptide sequence. Of these, 72 of the identified bacteriocin operons contained the full complement of accessory genes necessary for bacteriocin production but appeared to lack the required core structural peptides. Thirty-six strains encoded sactipeptide gene clusters with no core peptide, and 16 strains encoded bottromycin but again lacked the structural core peptide from 25 different genera including Actinomyces, Aerococcus, Bacillus, Fingoldia, Gordonia, Staphylococcus, Klebsiella, Leclercia, Morganella and Pseudomonas. A possible explanation for this is that the BAGEL4 database may simply not contain the sequence homologues of the core peptides[33]. It has also been hypothesised that bacteriocin production can be spontaneously acquired in the microbiome by horizontal gene transfer and can be lost by deletion of biosynthetic genes as bacteriocin production is metabolically costly[18,34]. Similarly, 20 putative helveticin J peptides predicted by BAGEL4 were excluded from further analysis due to the lack of a core peptide. It is also noteworthy that, in some studies, helveticin J peptides are no longer classed as bacteriocins and are considered a distinct group of antimicrobials (called bacteriolysins)[28].
Of the remaining 180 putative bacteriocin AOIs, 100 were determined to be lacking the key associated genes for bacteriocin production and, as such, were eliminated [Figure 1]. This resulted in 32 remaining isolates with 80 PBGCs that contained a structural core peptide and the associated accessory genes [Supplementary Table 2]. While they were removed based on the parameters set for this study, we do accept the possibility that these gene products may work in conjunction with other novel bacteriocins/bacteriocin-related genes encoded elsewhere on the genome[33]. Of the 80 remaining bacteriocins, 72% were identified as class II, 13.75% as class III, 8.75% as class I, and 5% as unclassified bacteriocins.
Further analysis of PBGCs of particular interest
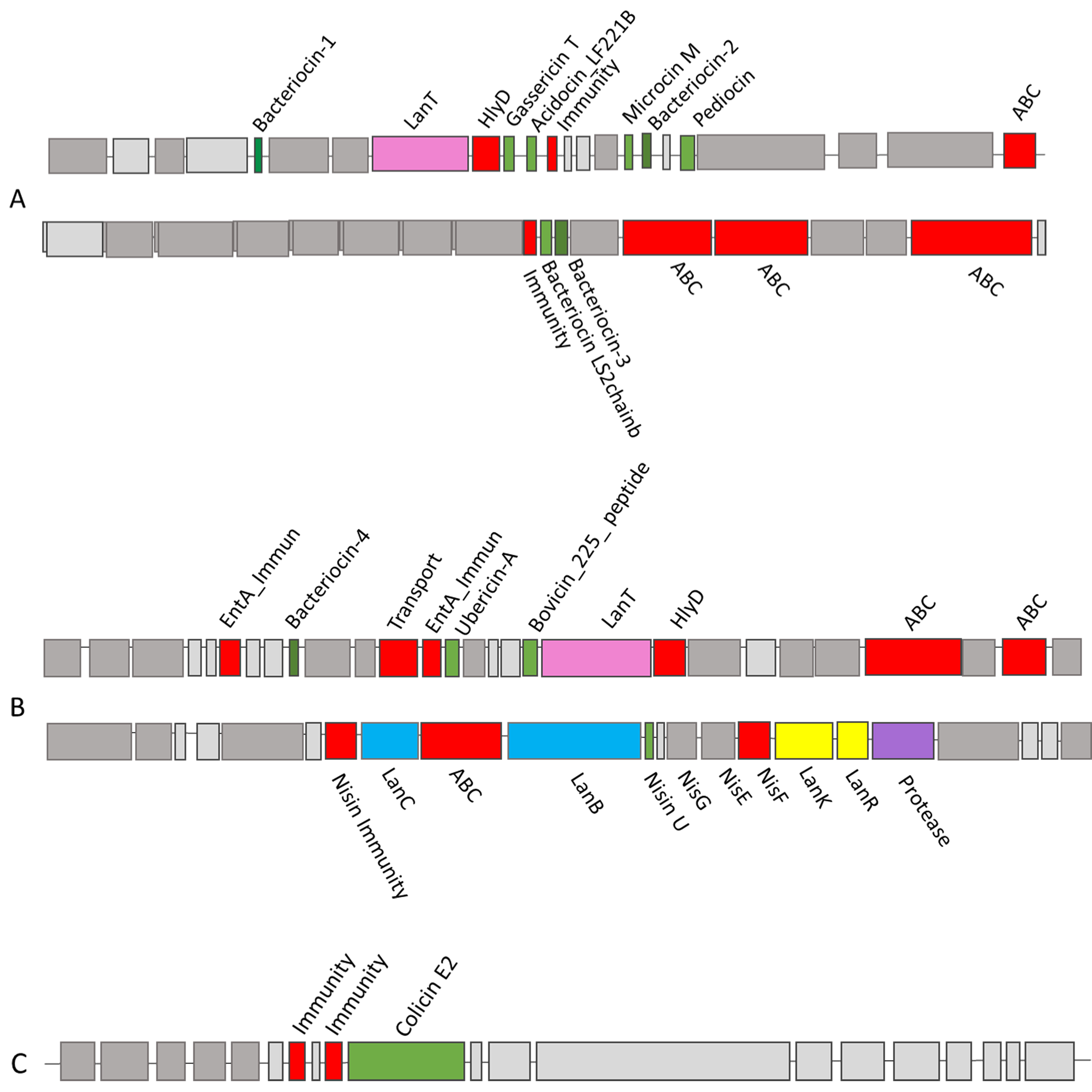
Based on BAGEL4, BLASTP, and EBI EMBOSS Needle analysis of the urobiome isolates, 53 putative bacteriocin hits were determined to be potentially novel [Supplementary Table 2]. Novelty in this case is taken as a core peptide with two or more amino acid differences compared to its closest characterised homologue[10], or a 100% identity to previously uncharacterised bacteriocin with no other closely related characterised homologues. Three bacterial strains, Lactobacillus gasseri UMB0099, Streptococcus macedonicus UMB0733, and Proteus mirabilis UMB0315, were chosen for further analysis [Table 1, Figures 2 and 3].
Figure 2. BAGEL4 outputs for urobiome isolates from (A) Lactobacillus gasseri UMB0099 depicting a putative microcin gene cluster and a class IIb gene cluster; (B) from Streptococcus macedonicus UMB0733 depicting a putative ubericin- A gene cluster and a putative novel nisin U gene cluster; and (C) from Proteus mirabilis UMB0315 depicting a putative active colicin bacteriocin gene cluster.
Figure 3. Mechanism of action of the putative bacteriocins identified in Table 1.
Abridged version of urobiome strains of interest which encode putative bacteriocin gene cluster(s) with surrounding accessory genes present
| BAGEL4 AOIs | Genome name | BLASTP result | Query cover | % identity | Alignment EMBOSS | Accession Number |
| ACIDOCIN_LF22IB (GASSERICINK7B) | Lactobacillus gasseri str. UMB0099 | Blp family class II bacteriocin [Lactobacillus] | 100% | 100% | 100% | WP_003649213.1 |
| BACTERIOCIN_LS2CHAINB* | Lactobacillus gasseri str. UMB0099 | Blp family class II bacteriocin [Ligilactobacillus salivarius] | 97% | 47.76% | 43.8% | WP_032495430.1 |
| BOVICIN_225_PEPTIDE* | Streptococcus macedonicus str. UMB0733 | Bacteriocin class II with double glycine leader peptide [Streptococcus infantarius subsp. Infantarius] | 100% | 93.5% | 95.9% | MCO4620482.1 |
| COLICIN* | Proteus mirabilis str. UMB0315 | Colicin-E2 [Proteus mirabilis] | 99% | 99% | 99.1% | AWF41001.1 |
| COMC; L_BIOTIC_TYPEA; BACTERIOCIN_IIC* | Streptococcus macedonicus str. UMB0733 | Bacteriocin [Streptococcus gallolyticus] | 100% | 78% | 78% | WP_114317773.1 |
| GASSERICIN_T | Lactobacillus gasseri str. UMB0099 | Blp family class II bacteriocin [Lactobacillus paragasseri] | 100% | 100% | 100% | WP_049159833.1 |
| GGMOTIF; BACTERIOCIN_IIC; | Lactobacillus gasseri str. UMB0099 | Blp family class II bacteriocin [Lactobacillus] | 100% | 100% | 100% | WP_101890487.1 |
| L_BIOTIC_TYPEA; BACTERIOCIN_IIC* | Lactobacillus gasseri str. UMB0099 | Lactacin F inducer peptide precursor [Lactobacillus johnsonii] | 100% | 84% | 92% | WP_260307981.1 |
| MICROCIN_M* | Lactobacillus gasseri str. UMB0099 | Bacteriocin [Lactobacillus] | 100% | 100% | 100% | WP_101890486.1 |
| NISIN_U* | Streptococcus macedonicus str. UMB0733 | Gallidermin/nisin family lantibiotic [Streptococcus suis] | 100% | 100% | 75% | WP_228478826.1 |
| PEDIOCIN* | Lactobacillus gasseri str. UMB0099 | Bacteriocin immunity protein [Lactobacillus] | 100% | 100% | 100% | WP_101890489.1 |
| UBERICIN_A* | Streptococcus macedonicus str. UMB0733 | Blp family class II bacteriocin [Streptococcus] | 93% | 100% | 93.5% | WP_003066580.1 |
| UNIDENTIFIED BACTERIOCIN* | Lactobacillus gasseri str. UMB0099 | Bacteriocin [Lactobacillus] | 100% | 100% | 100% | WP_049160225.1 |
Analysis of selected novel PBGCs identified among Lactobacillus species
Lactobacillus strains were identified most often (54%) when analysing urobiome isolates for novel bacteriocin production. Lactobacillus is one of the most common bacterial species isolated from the urobiome, particularly in women[13,28,35], and is thought to play a protective role within the urobiome[36]. Lactobacillus species are well-characterised bacteriocin producers[22] and have been highlighted for their potential applications in medicine, veterinary medicine, and the food industry as effective alternatives to antibiotics and food preservatives[37]. Additionally, many lactic acid bacteria (LAB) and their products are generally recognised as safe (GRAS status) by the FDA[37,38].
Lactobacillus gasseri UMB0099 was isolated by Miller-Ensminger et al. (2018)[30] from a urine sample collected by catheter from a patient suffering from an overactive bladder (OAB). BAGEL4 analysis revealed that this strain encoded nine putative bacteriocin peptides in total. One was removed as it lacked the associated production proteins. The remaining eight included three “generic” bacteriocins (dark green peptides; bacteriocin 1-3), gassericin T, acidocin_LF221B, microcin M, pediocin, and a bacteriocin LS2 chain b peptide [Figure 2A]. The gassericin T, acidocin_ LF221B (gassericin K7B), and bacteriocin-2 putative core peptides [Figure 2A] all represent 100% identity to previously characterised Blp class II bacteriocins[39,40]. Blp bacteriocin gene clusters are known to encode multiple bacteriocin-like peptides[39] which are secreted by ATP binding cassette (ABC) transporter proteins, similar to the PBGC identified in this study. Both gassericin and acidocin have been previously studied for their ability to prevent the growth of Staphylococcus aureus from mastitic milk and used as food preservatives and therapeutic agents for mastitis[41]. While the putative pediocin gene was predicted by BAGEL4 to encode a core bacteriocin peptide, once blasted, this was determined to exhibit 100% identity to an immunity protein and, thus, is likely mis-annotated and is not a core bacteriocin peptide. The putative cluster also contained a LanT gene, cleavage/export ABC transporter, HylD protein, and an immunity protein. The presence of all the accessory genes suggests functionality of the PBGC, although experimental validation is required for confirmation[41].
Of interest in this PBGC are the microcin M and bacteriocin-1. The microcin M on this cluster exhibits 100% identity to an uncharacterised bacteriocin, not a microcin. While BLASTP analysis did identify one uncharacterised bacteriocin, unlike other putative hits, it appears to have no other homologues. Microcins are typically produced by Gram-negative Enterobacteriaceae family[42] and have not been found before in Gram-positive bacteria. Therefore, with no other closely related hits, it can be inferred that it is either misannotated or is an uncharacterised novel bacteriocin. Bacteriocin-3 revealed a novel lactacin F precursor protein, with 84% identity to an uncharacterised lactacin F precursor isolated from Lactobacillus johnsonii. This is similar to other studies that have found multi-bacteriocin producing LAB strains that also encoded gassericin and acidocin on the same operon. These previously characterised bacteriocins have been shown to inhibit enteric bacteria and retain activity in colon conditions in vitro[43]. Production of multiple bacteriocins by a single strain is a desirable trait and can expand their spectrum of inhibition against different pathogenic strains. For example, Jiang et al. found synergistic activity, in vitro, of a two-peptide bacteriocin against a pathogenic Salmonella strain[44].
The putative class IIb PBGC shown in Figure 2A contained two core bacteriocin peptides: bacteriocin LS2 chain b and an unidentified bacteriocin. After analysis using BLASTP and EMBI-EBI EMBOSS Needle, the bacteriocin LS2 chain b showed a 43.8% identity to BIp family class II bacteriocin isolated from Ligilactobacillus salivarius[40], demonstrating a potentially novel variant of a class II bacteriocin. Similarly, the unidentified bacteriocin exhibited 100% identity to an uncharacterised bacteriocin; however, further investigation using BLASTP and EBI EMBOSS Needle alignment showed 91.1% identity to a previously characterised BIp family class II bacteriocin. These two putative BIp peptides were also found in Lactobacillus gasseri strain UMB0056. Among the Lactobacillus urobiome isolates, numerous strains encoded putative novel BIp class II bacteriocins with all the necessary accessory genes required for production present. Strains encoding novel class II two peptide bacteriocins include Lactobacillus crispatus strains UMB0040, UMB0803, UMB0805, UMB0044, UMB1398 and Lactobacillus rhamnosus UMB0004. Bacteriocins from LAB are of particular interest, with some already approved by the FDA for their use in food preservation, including nisin and pediocin[38]. Importantly, LAB bacteriocins have shown promise in inhibiting human pathogens in both in vitro studies against Pseudomonas aeruginosa[45], uropathogenic E.coli[46], and Candida spp.[47] and in in vivo studies against Gardnerella vaginalis[48], and in trials using bacteriocin-producing probiotics against bacterial vaginosis and UTIs[49]. Novel variants of these bacteriocins can aid in the treatment/prevention of infections, further highlighting the importance of in silico screening studies.
Analysis of various novel PBGCs identified from Streptococcus macedonicus
A study by Hilt et al. found Streptococcus to be a prevalent genus within the healthy female urobiome[50]. Streptococcus species produce many well-characterised bacteriocins[51] such as salivaricin, streptolysin, and mutacin[52], but none to date have been associated with the urobiome. Streptococcus macedonicus has previously been linked to bacteriocin production (macedocin) in dairy fermentations[53]. S. macedonicus UMB0733 was collected by catheter from a participant with no urinary symptoms or diseases[30]. BAGEL4 analysis on this strain identified multiple PBGCs.
The PBGC shown in Figure 2B was initially identified by BAGEL4 as an ubericin A bacteriocin gene cluster. Numerous Streptococcus uberis strains, producing multiple bacteriocins such as uberolysin A, ubericin A, and the lantibiotic, nisin U, have shown in vitro activity against different mastitis-inducing pathogens[54]. Further analysis of all three putative bacteriocin hits using BLASTP and EMBI-EBI EMBOSS Needle identified novel variants of class II bacteriocin peptides. The first bacteriocin hit shared 78% identity with an uncharacterised bacteriocin isolated from Streptococcus gallolyticus. The ubericin A hit shared 93.5% identity with a BIp family class II bacteriocin previously characterised by Dawid et al.[39].
Finally, the bovicin 225 peptide shared 93.4% identity with a previously characterised class II bacteriocin with a double-glycine leader peptide isolated from Streptococcus infantarius. Alongside the three core peptides, two EntA immunity proteins were present on the genome with LanT, HlyD, and two ABC transporters. With three putative novel bacteriocins and all the accessory genes necessary for production, it can be inferred that this is a novel variant of class II bacteriocin gene cluster. Figure 2B shows a novel variant of the bacteriocin nisin U which displayed 75% identity to a gallidermin/nisin family lantibiotic previously characterised by Christ et al.[55]. With all eleven accessory genes present on the gene cluster [Figure 2B], it can be assumed to be an active variant of nisin U[56].
Analysis of novel Colicin PBGCs identified from Proteus mirabilis
Proteus mirabilis UMB0315 [Figure 2C] was collected by catheter from a participant with symptoms of an overactive bladder[30]. BAGEL4 analysis identified a novel colicin bacteriocin gene cluster. Colicins, which are usually produced by E. coli, are among the most well-studied bacteriocins and are effective antimicrobials against other E. coli and Enterobacteriaceae strains[57,58]. The colicin of interest in this study was a novel variant produced by Proteus mirabilis. To date, it appears that colicins produced by Proteus have not been extensively characterised, but a crude bacteriocin extract from Proteus mirabilis has been described for its colicin-like antibiofilm properties[59].
The PBGC, shown in Figure 2C, was identified by BAGEL4 as a colicin E2 bacteriocin gene cluster. Further analysis of the bacteriocin core peptide using BLASTP and EMBI-EBI EMBOSS demonstrated a novel variant of a colicin E2 peptide sharing 99.5% identity with six amino acid differences at positions 1, 57, 59, 86, 128, and 188. With both immunity proteins also present [Figure 2C], this suggests that this novel colicin variant is one of the first predicted to be produced by a Proteus species. Colicins have been previously highlighted for their antimicrobial potential by coating catheters to inhibit colonisation by UTI-causing pathogenic bacteria[60]. Colicins have numerous favourable properties such as the low concentrations needed for antimicrobial activity and also their specificity in killing, making them desirable antimicrobials that inflict limited collateral damage on the commensal microbiota[61]. Other putative colicin clusters identified in this study were found among the following urobiome strains: Citrobacter murliniae (UMB1094), Escherichia fergusonii (UMB0727, UMB0901, UMB0900, UMB0789), Morganella morganii (UMB1297) and Pseudomonas aeruginosa (UMB0710). New bacteriocins that have recently obtained GRAS status from the FDA for their use in food include colicins and colicin-like peptides (salmocins from Salmonella)[58], further highlighting the utility of in silico screening studies to aid in the discovery of novel bacteriocins.
DISCUSSION
In conclusion, previous in silico screening techniques have successfully identified bacteriocin gene clusters in the human microbiome[5,6,9-11,33]. However, given that urobiome research is in its nascent stages, investigation of its bioactive products has remained relatively understudied to date. This in silico analysis highlights the overall bacteriocin production ability of the urobiome. Bacteriocin production is a highly regulated process and requires specific environmental conditions, which complicates in vitro screening for bacteriocins. In silico screening, on the other hand, has allowed the rapid identification of bacteriocins without the restrictions of in vitro screening[10,28]. However, it is important to note that in silico screening is limited by the need for comparison to previously characterised bacteriocins, which can lead to completely novel bacteriocin gene clusters being missed[28,33]. Furthermore, such in silico screens are based solely on inference and can only be definitively verified by in vitro and or in vivo follow-up analysis. Notwithstanding, the current study determined that 19.33% (35/181) of strains isolated from the urobiome encoded one or more potentially active bacteriocin peptides. Despite these limitations, such studies remain an important first step in identifying novel bacteriocins. Bacteriocins have been isolated from a variety of microbiomes, demonstrating antimicrobial activity against clinically relevant pathogens. Bacteriocins have been used to target pathogens both in vitro[45-47,54,62] and in vivo[48,63], exerting probiotic effects[49,64], inhibiting biofilm formation[60], and resensitising resistant bacterial strains to antibiotics[65], while demonstrating limited cytotoxic effects on the commensal healthy microbiomes. It is hoped that the identification of bacteriocins from untapped niches such as the urobiome can aid in the transition into use in clinical settings to control infections.
Overall, this study identified 53 putative novel bacteriocin peptides that have not been previously characterised, suggesting a high degree of novelty and diversity of bacteriocin production within the urobiome. These results indicate that the urobiome represents a comprehensive, and relatively untapped, source of novel antimicrobials, some of which might well find application in the control of antibiotic resistant infections.
DECLARATIONS
Authors’ contributions
Literature search: Culligan EP, Jones J
Data collection and analysis: Jones J
Writing and editing: Jones J
Review and editing: Culligan EP, Sleator RD, Murphy CP
Supervision of research project: Culligan EP, Sleator RD, Murphy CP
Availability of data and materials
The accession numbers for all genomes used for analysis in this manuscript can be found in Supplementary Data. All BAGLE4 and antiSMASH data can be found in Supplementary Tables 1 and 2.
Financial support and sponsorship
This research was funded by an MTU RISAM PhD Fellowship to Jones J.
Conflicts of interest
All authors declared that there are no conflicts of interest.
Ethical approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Copyright
© The Author(s) 2024.
Supplementary Materials
REFERENCES
1. Aragón IM, Herrera-Imbroda B, Queipo-Ortuño MI, et al. The urinary tract microbiome in health and disease. Eur Urol Focus 2018;4:128-38.
2. Newstead LL, Varjonen K, Nuttall T, Paterson GK. Staphylococcal-produced bacteriocins and antimicrobial peptides: their potential as alternative treatments for Staphylococcus aureus infections. Antibiotics 2020;9:40.
3. Valles-Colomer M, Manghi P, Cumbo F, et al. Neuroblastoma is associated with alterations in gut microbiome composition subsequent to maternal microbial seeding. EBioMedicine 2024;99:104917.
4. Xie J, Liu M, Deng X, et al. Gut microbiota reshapes cancer immunotherapy efficacy: mechanisms and therapeutic strategies. iMeta 2024;3:e156.
5. Zheng J, Gänzle MG, Lin XB, Ruan L, Sun M. Diversity and dynamics of bacteriocins from human microbiome. Environ Microbiol 2015;17:2133-43.
6. Mousa WK, Athar B, Merwin NJ, Magarvey NA. Antibiotics and specialized metabolites from the human microbiota. Nat Prod Rep 2017;34:1302-31.
7. Culligan EP, Sleator RD. Advances in the microbiome: applications to Clostridium difficile infection. J Clin Med 2016;5:83.
8. O’Sullivan JN, Rea MC, O’Connor PM, Hill C, Ross RP. Human skin microbiota is a rich source of bacteriocin-producing staphylococci that kill human pathogens. FEMS Microbiol Ecol 2019;95:fiy241.
9. Wosinska L, Walsh CJ, O’Connor PM, et al. In vitro and in silico based approaches to identify potential novel bacteriocins from the athlete gut microbiome of an elite athlete cohort. Microorganisms 2022;10:701.
10. Angelopoulou A, Warda AK, O’Connor PM, et al. Diverse bacteriocins produced by strains from the human milk microbiota. Front Microbiol 2020;11:788.
11. King AM, Zhang Z, Glassey E, Siuti P, Clardy J, Voigt CA. Systematic mining of the human microbiome identifies antimicrobial peptides with diverse activity spectra. Nat Microbiol 2023;8:2420-34.
12. Jones J, Murphy CP, Sleator RD, Culligan EP. The urobiome, urinary tract infections, and the need for alternative therapeutics. Microb Pathog 2021;161:105295.
14. Acedo JZ, Chiorean S, Vederas JC, van Belkum MJ. The expanding structural variety among bacteriocins from Gram-positive bacteria. FEMS Microbiol Rev 2018;42:805-28.
15. Chikindas ML, Weeks R, Drider D, Chistyakov VA, Dicks LM. Functions and emerging applications of bacteriocins. Curr Opin Biotechnol 2018;49:23-8.
16. Riley MA, Wertz JE. Bacteriocins: evolution, ecology, and application. Annu Rev Microbiol 2002;56:117-37.
17. Héchard Y, Sahl HG. Mode of action of modified and unmodified bacteriocins from Gram-positive bacteria. Biochimie 2002;84:545-57.
18. Heilbronner S, Krismer B, Brötz-Oesterhelt H, Peschel A. The microbiome-shaping roles of bacteriocins. Nat Rev Microbiol 2021;19:726-39.
19. Chavan MA, Riley MA. Molecular evolution of bacteriocins in gram-negative bacteria. In: Riley MA, Chavan MA, editors. Bacteriocins. Berlin: Springer Berlin Heidelberg; 2007. pp. 19-43.
20. Ghodhbane H, Elaidi S, Sabatier JM, Achour S, Benhmida J, Regaya I. Bacteriocins active against multi-resistant gram negative bacteria implicated in nosocomial infections. Infect Disord Drug Targets 2015;15:2-12.
23. Cotter PD, Ross RP, Hill C. Bacteriocins - a viable alternative to antibiotics? Nat Rev Microbiol 2013;11:95-105.
24. Benítez-Chao DF, León-Buitimea A, Lerma-Escalera JA, Morones-Ramírez JR. Bacteriocins: an overview of antimicrobial, toxicity, and biosafety assessment by in vivo models. Front Microbiol 2021;12:630695.
25. Egan K, Field D, Ross RP, Cotter PD, Hill C. In silico prediction and exploration of potential bacteriocin gene clusters within the bacterial genus Geobacillus. Front Microbiol 2018;9:2116.
26. van Heel AJ, de Jong A, Song C, Viel JH, Kok J, Kuipers OP. BAGEL4: a user-friendly web server to thoroughly mine RiPPs and bacteriocins. Nucleic Acids Res 2018;46:W278-81.
27. Blin K, Shaw S, Augustijn HE, et al. antiSMASH 7.0: new and improved predictions for detection, regulation, chemical structures and visualisation. Nucleic Acids Res 2023;51:W46-50.
28. Collins FWJ, O’Connor PM, O’Sullivan O, et al. Bacteriocin gene-trait matching across the complete Lactobacillus pan-genome. Sci Rep 2017;7:3481.
29. Conrads G, Westenberger J, Lürkens M, Abdelbary MMH. Isolation and bacteriocin-related typing of Streptococcus dentisani. Front Cell Infect Microbiol 2019;9:110.
30. Miller-Ensminger T, Garretto A, Brenner J, et al. Bacteriophages of the urinary microbiome. J Bacteriol 2018;200:e00738-17.
31. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol 1990;215:403-10.
32. Brubaker L, Gourdine JPF, Siddiqui NY, et al. Forming consensus to advance urobiome research. mSystems 2021;6:e0137120.
33. Walsh CJ, Guinane CM, Hill C, Ross RP, O’Toole PW, Cotter PD. In silico identification of bacteriocin gene clusters in the gastrointestinal tract, based on the Human Microbiome Project’s reference genome database. BMC Microbiol 2015;15:183.
34. Christenson JK, Gordon DM. Evolution of colicin BM plasmids: the loss of the colicin B activity gene. Microbiology 2009;155:1645-55.
35. Wolfe AJ, Brubaker L. Urobiome updates: advances in urinary microbiome research. Nat Rev Urol 2019;16:73-4.
36. Roth RS, Liden M, Huttner A. The urobiome in men and women: a clinical review. Clin Microbiol Infect 2023;29:1242-8.
37. Daba GM, Elkhateeb WA. Bacteriocins of lactic acid bacteria as biotechnological tools in food and pharmaceuticals: current applications and future prospects. Biocatal Agric Biotechnol 2020;28:101750.
38. Mokoena MP. Lactic acid bacteria and their bacteriocins: classification, biosynthesis and applications against uropathogens: a mini-review. Molecules 2017;22:1255.
39. Dawid S, Roche AM, Weiser JN. The blp bacteriocins of Streptococcus pneumoniae mediate intraspecies competition both in vitro and in vivo. Infect Immun 2007;75:443-51.
40. Franz CMAP, Van Belkum MJ, Holzapfel WH, Abriouel H, Gálvez A. Diversity of enterococcal bacteriocins and their grouping in a new classification scheme. FEMS Microbiol Rev 2007;31:293-310.
41. Kasuga G, Tanaka M, Harada Y, et al. Homologous expression and characterization of gassericin T and gassericin S, a novel class IIb bacteriocin produced by Lactobacillus gasseri LA327. Appl Environ Microbiol 2019;85:e02815-18.
42. Vassiliadis G, Destoumieux-Garzón D, Lombard C, Rebuffat S, Peduzzi J. Isolation and characterization of two members of the siderophore-microcin family, microcins M and H47. Antimicrob Agents Chemother 2010;54:288-97.
43. Garcia-Gutierrez E, O’Connor PM, Colquhoun IJ, et al. Production of multiple bacteriocins, including the novel bacteriocin gassericin M, by Lactobacillus gasseri LM19, a strain isolated from human milk. Appl Microbiol Biotechnol 2020;104:3869-84.
44. Jiang H, Li P, Gu Q. Heterologous expression and purification of plantaricin NC8, a two-peptide bacteriocin against Salmonella spp. from Lactobacillus plantarum ZJ316. Protein Expr Purif 2016;127:28-34.
45. Jamalifar H, Rahimi HR, Samadi N, et al. Antimicrobial activity of different Lactobacillus species against multi-drug resistant clinical isolates of Pseudomonas aeruginosa. Iran J Microbiol 2011;3:21-5.
46. Butler DSC, Silvestroni A, Stapleton AE. Cytoprotective effect of Lactobacillus crispatus CTV-05 against Uropathogenic E. coli. Pathogens 2016;5:27.
47. Reid G, Bruce AW, Fraser N, Heinemann C, Owen J, Henning B. Oral probiotics can resolve urogenital infections. FEMS Immunol Med Microbiol 2001;30:49-52.
48. Ya W, Reifer C, Miller LE. Efficacy of vaginal probiotic capsules for recurrent bacterial vaginosis: a double-blind, randomized, placebo-controlled study. Am J Obstet Gynecol 2010;203:120.e1-6.
49. Beyitler I, Kavukcu S. Probiotics for prophylaxis and treatment of urinary tract infections in children. Iran J Pediatr 2016;27:e7695.
50. Hilt EE, McKinley K, Pearce MM, et al. Urine is not sterile: use of enhanced urine culture techniques to detect resident bacterial flora in the adult female bladder. J Clin Microbiol 2014;52:871-6.
51. Tagg JR, Dajani AS, Wannamaker LW. Bacteriocins of gram-positive bacteria. Bacteriol Rev 1976;40:722-56.
52. Nes IF, Diep DB, Holo H. Bacteriocin diversity in Streptococcus and Enterococcus. J Bacteriol 2007;189:1189-98.
53. De Vuyst L, Tsakalidou E. Streptococcus macedonicus, a multi-functional and promising species for dairy fermentations. Int Dairy J 2008;18:476-85.
54. Lasagno M, Navarro MLA, Moliva M, Reinoso E. Screening of bacteriocin associated genes of Streptococcus uberis strains. Heliyon 2019;5:e02393.
55. Christ K, Al-Kaddah S, Wiedemann I, Rattay B, Sahl HG, Bendas G. Membrane lipids determine the antibiotic activity of the lantibiotic gallidermin. J Membr Biol 2008;226:9-16.
56. Wirawan RE, Klesse NA, Jack RW, Tagg JR. Molecular and genetic characterization of a novel nisin variant produced by Streptococcus uberis. Appl Environ Microbiol 2006;72:1148-56.
57. Marković KG, Grujović MŽ, Koraćević MG, et al. Colicins and microcins produced by Enterobacteriaceae: characterization, mode of action, and putative applications. Int J Environ Res Public Health 2022;19:11825.
58. Hahn-Löbmann S, Stephan A, Schulz S, et al. Colicins and salmocins - new classes of plant-made non-antibiotic food antibacterials. Front Plant Sci 2019;10:437.
59. Hussein AR, Khalaf ZZ, Kadhim MJ. The antibiofilm activity of bacteriocin produced by Proteus mirabilis against some bacterial species. Curr Res Microbiol Biotechnol 2017;5:1071-7. Available from: https://www.researchgate.net/profile/Alyaa-Hussein-2/publication/328052134_The_antibiofilm_activity_of_bacteriocin_produced_by_Proteus_mirabilis_against_some_bacterial_species/links/5bb5208ca6fdccd3cb8517fd/The-antibiofilm-activity-of-bacteriocin-produced-by-Proteus-mirabilis-against-some-bacterial-species.pdf. [Last accessed on 26 Mar 2024]
60. Roy SM, Riley MA. Evaluation of the potential of colicins to prevent extraluminal contamination of urinary catheters by Escherichia coli. Int J Antimicrob Agents 2019;54:619-25.
61. Sharp C, Boinett C, Cain A, et al. O-antigen-dependent colicin insensitivity of uropathogenic Escherichia coli. J Bacteriol 2019;201:e00545-18.
62. Sharma D, Gajjar D, Seshadri S. Understanding the role of gut microfloral bifidobacterium in cancer and its potential therapeutic applications. Microbiome Res Rep 2024;3:3.
63. Sassone-Corsi M, Nuccio SP, Liu H, et al. Microcins mediate competition among Enterobacteriaceae in the inflamed gut. Nature 2016;540:280-3.
64. Culligan EP, Hill C, Sleator RD. Probiotics and gastrointestinal disease: successes, problems and future prospects. Gut Pathog 2009;1:19.
Cite This Article
Export citation file: BibTeX | RIS
OAE Style
Jones J, Murphy CP, Sleator RD, Culligan EP. An exploratory in silico analysis of bacteriocin gene clusters in the urobiome. Microbiome Res Rep 2024;3:24. http://dx.doi.org/10.20517/mrr.2023.78
AMA Style
Jones J, Murphy CP, Sleator RD, Culligan EP. An exploratory in silico analysis of bacteriocin gene clusters in the urobiome. Microbiome Research Reports. 2024; 3(2): 24. http://dx.doi.org/10.20517/mrr.2023.78
Chicago/Turabian Style
Jones, Jennifer, Craig P. Murphy, Roy D. Sleator, Eamonn P. Culligan. 2024. "An exploratory in silico analysis of bacteriocin gene clusters in the urobiome" Microbiome Research Reports. 3, no.2: 24. http://dx.doi.org/10.20517/mrr.2023.78
ACS Style
Jones, J.; Murphy CP.; Sleator RD.; Culligan EP. An exploratory in silico analysis of bacteriocin gene clusters in the urobiome. Microbiome. Res. Rep. 2024, 3, 24. http://dx.doi.org/10.20517/mrr.2023.78
About This Article
Special Issue
Copyright
Data & Comments
Data

 Cite This Article 1 clicks
Cite This Article 1 clicks











Comments
Comments must be written in English. Spam, offensive content, impersonation, and private information will not be permitted. If any comment is reported and identified as inappropriate content by OAE staff, the comment will be removed without notice. If you have any queries or need any help, please contact us at support@oaepublish.com.